HMGI(Y)
From Proteopedia
June Hope 20:21, 3 June 2014 (IDT)
|
Contents |
Structure
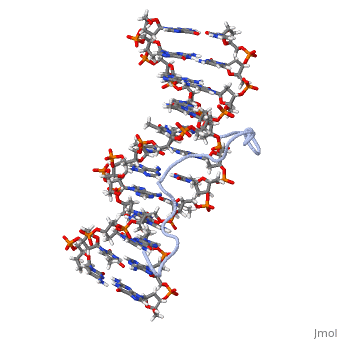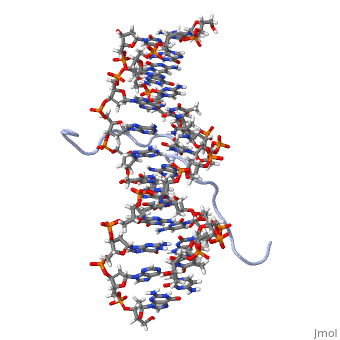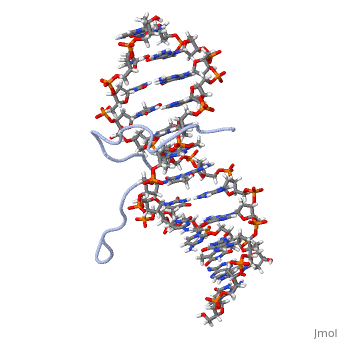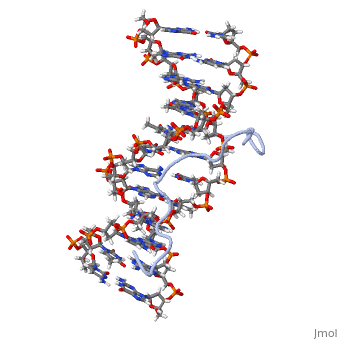
High Mobility Group (HMG) proteins are a class of non-histone chromosomal proteins found in all mammals. HMG proteins recognize and bind DNA and are involved in the regulation of various DNA-dependent processes, such as transcription, recombination and DNA repair. HMGI(Y) is a High Mobility Group (HMG) protein with two isomers—I and Y. These proteins were isolated from human cells in 1983, and were of particular interest because they exhibit preferential binding to the minor groove of AT rich regions of double-stranded DNA, which is essential to many of its functions. HMGI(Y) proteins have three DNA binding sites, termed AT hook DNA-binding domains, found in discrete locations between amino acids 21 and 89. Overlapping two of the AT hook sites is a region that specifies in interacting with HIPK2. There is also a glutamic acid-rich (acidic) region located at one end of the protein, amino acids 91-106.
HMGI(Y) Sequence Length: 107 AA
Molecular Mass: 11750-11858 Da
PDB: P17096 [1]
Function of HMGI(Y)
The High-mobility group protein HMGI(Y) is a mammalian non-histone chromosomal protein that participates in a variety of cellular processes. HMGI(Y) is involved in regulation of inducible gene transcription and structural changes in chromatin substrates and other proteins.
Among other cellular processes, in a healthy human being, HMGI(Y) plays a crucial role in cytokine gene transcription in CD4+ T-helper cells. CD4+ cells have the ability to differentiate into two different phenotypes, varying in their cytokine profile so that they have the ability to elicit a more specific immune response to an infection. In order to do this, HMGI(Y) modulates the function of several transcription factors that in turn control the cytokine promoter, thus HMGI(Y) determines the ratio of the two different CD4+ phenotypes produced, allowing for more specific immune response activities.
Associated Disease: HIV-1
The Human Immunodeficiency Virus (HIV) is a retrovirus that causes the condition known as acquired immunodeficiency syndrome, or AIDs. HIV is a commonly known sexually transmitted disease, which weakens the body’s immune system by taking over control of host cell processes. The HIV virus attacks and destroys CD4+ T-helper cells—immune system cells—by infecting the cell and then using the host cell’s genetic material and proteins to replicate itself. Replication and subsequent release of new virus particles kill CD4+ cells, and weaken the immune system of the individual.
Function of HMGI(Y) in HIV
In a person infected with HIV-1, HMGI(Y) is required for function of HIV-1 pre-integration complexes (PICs). PICs are complexes that contain viral genetic material, along with associated viral and host proteins, and which are capable of inserting a viral genome into a host genome. Though the exact mechanism is still unknown, it has been shown that HMGI(Y) is required for HIV-1 cDNA integration into PICs. Current proposed mechanisms include HMGI(Y) binding to either viral cDNA and creating a necessary conformational change in the cDNA or HMGI(Y) binding to integrase and acting as an allosteric regulator. Due to the AT hook DNA-binding domains, it is likely that HMGI(Y) binds to cDNA.
Therapeutic Agents Used to Target HMGI(Y)
Since the mechanism of HMGI(Y) is unknown, there are no current therapeutic agents available for targeting this specific protein. Possible future therapeutic agents could potentially target the active site of HMGI(Y) which recognizes and binds DNA, or bind to areas that cause a conformational change in HMGI(Y) making it unable to bind to other proteins or DNA.
Current therapeutic agents that don’t target HMGI(Y) directly but affect its activity are agents that target HIV integrase.
3D Structures of high mobility group protein (HMG)
See High mobility group protein
References
http://www.rcsb.org/pdb/results/results.do?outformat=&qrid=369566F9&tabtoshow=Current
http://www.nature.com/icb/journal/v76/n5/full/icb199862a.html
http://www.nature.com/icb/journal/v76/n5/pdf/icb199862a.pdf
http://www.sciencedirect.com/science/article/pii/S0092867400818887
http://www.retrovirology.com/content/7/1/66
http://cid.oxfordjournals.org/content/41/Supplement_1/S96.long