We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
From Proteopedia
proteopedia linkproteopedia link Malate Dehydrogenase
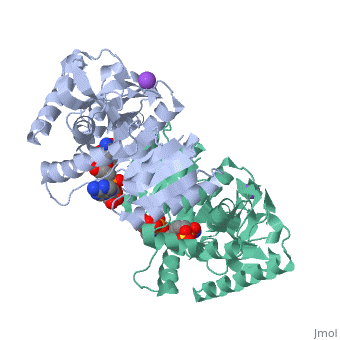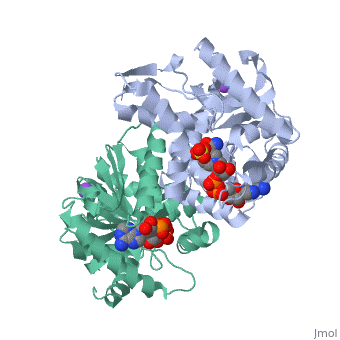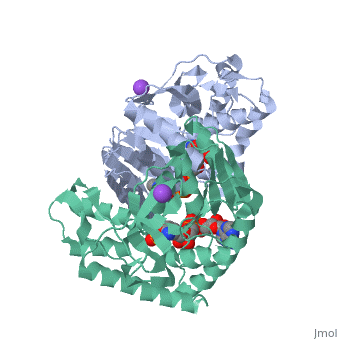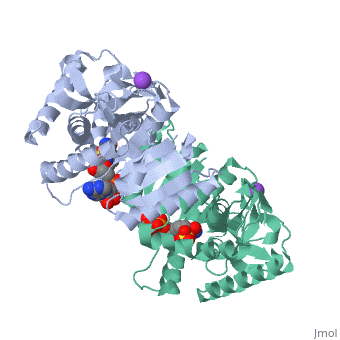
Malate Dehydrogenase (MDH)(PDB entry 2x0i) is most known for its role in the metabolic pathway in the tricarboxylic acid cycle[1]; however, the enzyme is also in many other metabolic pathways such as glyoxylate bypass, amino acid synthesis, glucogenesis, and oxidation/reduction balance [1]. It is classified as a oxidoreductase[2]. Malate Dehydrogenase has been extensively studied due to its many isozymes [2]. The enzyme exists in two places inside a cell, in the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; but in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport [3]. This conversion is an essential catalytic step in each different metabolic mechanism. The enzyme malate dehydrogenase is composed of either a dimer or tetramer [4].
The evolutionary past of MDH shows a divergence to form lactate dehydrogenase (LDH) which functions in a very similar way to MDH. Although there is a very low sequence conservation among MDH and LDH’s [3] the structure of the enzyme has remained relatively conserved. The key difference between the two is in the substrate: LDH catalyzes pyruvate to lactate.
|
|
| 2x0i, resolution 2.91Å ()
|
| Ligands:
| , ,
|
| Activity:
| Malate dehydrogenase, with EC number 1.1.1.37
|
| Related:
| 2x06, 2x0j, 2x0n, 2x0r, 2x0s
|
|
|
|
|
| Resources:
| FirstGlance, OCA, RCSB, PDBsum
|
| Coordinates:
| save as pdb, mmCIF, xml
|