Molecular Playground/T7 RNAP Conformations
From Proteopedia
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground
| ||||||
|
Color code:
|
Contents |
Conformational Changes on T7 RNA Polymerase
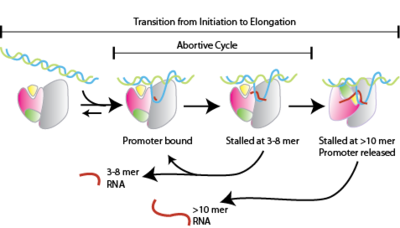
Transcription is a fundamental part of genetic regulation. The RNA polymerases that accomplish this function vary in structure, size and complexity, but must all carry out the same basic functions ([See[1]RNA polymerases). The correct transcription of DNA to RNA depends of several factors and the complexity increases with the complexity of the organism. This makes the study of the transcriptional process complicated. The RNA polymerase of the bacteriophage T7, is the perfect model for studying the transcription process given that T7 RNA polymerase is a single unit enzyme that processes RNA with the same effectivity as the polymerase from higher organisms. Nevertheless, there is plenty to learn from the transcription mechanism, such as the "abortive cycle" process that takes place during the phase (Figure 1) remains poorly understood.
In this event the small RNA transcripts (less than 12 bases) dissociate from the complex. The abortive cycle will continue until the enzyme/DNA/RNA complex reaches the phase in order to for a more stable enzyme/DNA/RNA complex. A mayor contributor of the stability of the complex is the formation of the . Another interesting observation that could help to resolve the mechanism of abortive cycling, is a single point mutation at the . This mutation is far away from the and the region and it is located on the hinge between the N-terminus and the C-terminus. Although leucine is not the only substitution that decreases the amount of abortive products, it is the one with the mayor effect. It is proposed that the mutation creates a more flexible protein structure that facilitates the transition from initiation to elongation (notice the position of the ). Part of our research is focused on resolving the mechanism behind this mutation.
Understanding the Morph
In order to activate the transition between the conformation and the complex, click the following button.
The first striking observation is the conformational change of the N-terminus part of the enzyme and the helices C1-C2. The DNA with translucent colors is our reference point and the modeled DNA is part of the intermediate state structure. The N-terminus rotates around 47º, the RNA transcript has 7 bases, but the enzyme has not reached its final elongation conformation yet. The missing steps could be resolved if we morph the structures using the intermediate state and the elongation structures. The following shows a complete refolding of the sub-domain H (alfa-helices in green) and the helices C-1 C-2. It uses the intermediate state and the elongation state. However, there is a problem. Can you see it? Follow the movement of the green helices. Indeed, it can not be the real transition. While there has been good advances in solving the correct transition (2), the optimal way is by producing structures of the transitional complexes from 9 and 10 mer transcripts. Another approach to study this transition would be by labeling the enzyme with fluorophores and then using FRET, which could allow us to calculate the movement distances that occurs during the transition. This work is in progress... Finally, the morphs were produced using the energy minimization morphing software from the Yale Morph Server. The protein structures that were used in the server are the following: T7 RNA polymerase initiation complex (PDB ID:1qln), T7 intermediate state complex (PDB ID:3e2e)(1) and the T7 RNA polymerase elongation complex (PDB ID:1msw).
3D structures of RNA polymerase
References
- Steitz, T. A. (2009) The structural changes of T7 RNA polymerase from transcription initiation to elongation., Curr. Opin. Struct. Biol. 19, 683-690.
- Turingan, R. S., Theis, K., and Martin, C. T. (2007) Twisted or shifted? Fluorescence measurements of late intermediates in transcription initiation by T7 RNA polymerase., Biochemistry 46, 6165-6168.
Acknowledgement
To Professor Eric Martz his advice was crucial to develop this page.