p53 Tumor Suppressor Protein (PDB ID 1tup)
p53 is a tumor suppressor. In the absence of cellular stress, p53 does not exert effect on cell fate, but under stress, p53 becomes activated and causes phenotypic changes in cells like senescence and apoptosis[1]. Many human cancer cells carry mutated p53[2]. The name p53 refers to its apparent molecular mass. It runs as a 53 kDa molecule on SDS-PAGE. But based on calculations from its amino acid residues, p53's mass is actually 43.7 kDa. This difference may be due to the high number of proline residues in the protein, resulting in its migrating slowly on SDS-PAGE.
Human p53 is 393 amino acids long and has seven domains:
- Transcription activation domain (TAD residues 15-29)
- Activation domain 2
- Proline rich domain
- DNA-binding core domain (DBD residues 94-292)
- A nuclear localization signaling domain
- Tetramerizatin domain (TD residues 326-356)
- C-terminal domain
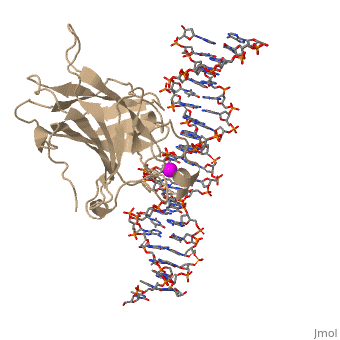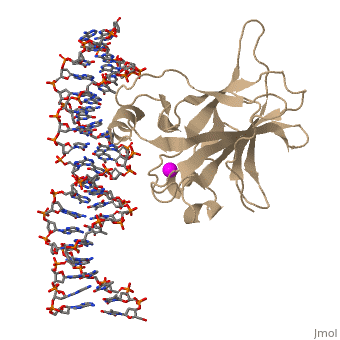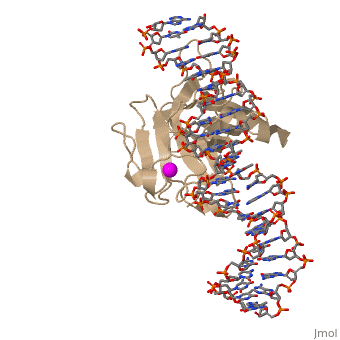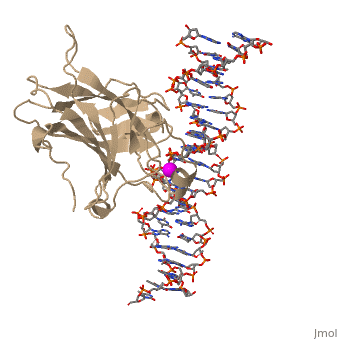
p53 tumor suppressor is a flexible molecule composed of four identical protein chains. Flexible molecules are difficult to study by X-ray crystallography because they do not form orderly crystals. So p53 has been studied in parts, by removing the flexible regions and solving structures of the pieces that form stable structures. The figure at the right shows the cartoon representation of the DNA-binding domain, which has been studied most.
For more details see P53-DNA Recognition
Oncogenes & Tumor Suppressor Genes
See also P53 (hebrew).
p53 Pathway and Mutation
In a normal cell, p53 is inactivated by its negative regulatory mdm2 (hdm2 in humans) and it is found at low levels. When DNA damage is sensed, p53's level rises. p53 binds to many regulatory sites in the genome and begins production of proteins that stop cell division until the damage is repaired. If the damage is irreparable, p53 initiates the process called programmed cell death, apoptosis, permanently removing the damage.
In most cases of human cancer, p53 mutations have been observed. Most of the p53 mutations that may result in cancer are found in and around the DNA-binding surface of the protein. The most common mutation changes can be seen in a close up view of the color coded N to C (in rainbow colors), with the amino (as space filling spheres) interacting with DNA. When mutated to another amino acid, this interaction is lost. Other key residues associated with cancer-causing mutations are represented by magenta spheres.
Surface charge of the DNA binding domain
The figure at the left shows the surface charge of the p53 DNA-binding domain )Charged: red - negative; blue - positive). It is rich in arginine amino acids that interact with DNA, and this causes its surface to be positively charged. This domain recognizes specific regulatory sites on the DNA. The flexible structure of p53 allows it to bind to many different variants of binding sites, allowing it to regulate transcription at many places in the genome.
There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues that are located on two loops, respectively. It is conceivable that the zinc plays a role in stabilizing the two loops through
coordination. The Zn has been represented as a magenta sphere, and R248 in space filling, in the scene at the right.
3D structures of p53
P53 3D structures