STRUCTURE 1BM0
From Proteopedia
|
Crystal structure of human serum albumin at 2.5 A°
Members of CSUCI BIOL 505 Molecular Structure Project: Neeharika Pothuri, Bhagiradhi Somalanka, Irene Becerra, Valerie Ajiduah
|
| About • Editing • Help | Video Guide • Table of Contents • Content (Topic Pages) • What's New |
Contents |
Abstract
The three dimensional structures of pHSA and rHSA triclinic crystals were determined at 2.5A°resolution through multiple isomorphous replacement with a known tetragonal crystal. The pHSA and rHSA structures are similar with an r.m.s deviation of 0.24 A° for all the cα atoms. Cys 34 does not have a disulfide link with any ligand. A pocket of hydrophobic and positively charged residues is formed at domains 2 and 3 where a wide range of compounds can be accommodated. The surface of the domain has three binding sites for long chain fatty acids.
Introduction
HSA is the most abundant protein in the blood plasma at a blood concentration of 5g/100ml. HSA has a high affinity for Cu+2, Zn+2, fatty acids, amino acids, metabolites and many other drug compounds. The protein brings solutes in the blood to the target organs and also maintains the pH and osmotic pressure of the plasma hence it is used as a carrier of drugs to their targets. HSA has 585 residues with 17 disulfide bridges and one free cysteine. Many reports contained crystal forms of HSA but none provided structural information. This article discusses the crystal form of defatted HSA and the molecular aspects of the protein are determined at the highest resolution.
The image on the right shows a representation of HSA structure (in ribbon display) with myristate, azidothymidine (AZT), and salicylic acid in van der Waals dots.
Crystallization and structural determination of the tetragonal crystal
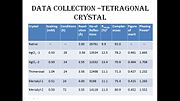
pHSA purchased from sigma chemical co. was crystallized using the hanging drop method. Yellow colored crystals were obtained from a solution containing 150-255 mg/ml protein, 50mM potassium phosphate, 30-38% PEG 400 and 5mM sodium azide. The crystals belonged to tetragonal space group P4212 with cell dimensions a=187.1A°, c=80.5A°. Crystals reach a resolution of 3.0A° due to high solvent concentration. Intensity data was collected using Rigaku R-AXIS 11c area detector on a rigaku RU-200H x-ray generator. The diffraction data was processed with PROCESS. Hgcl2 was used for phasing with multiple isomorphous replacement. The heavy atom sites were located using the HASSP program. The phasing statistics can be seen in table 1.
Crystallization and structural determination of the triclinic crystal
Before crystallization fatty acids were removed from the sample solution using activated powdered charcoal. The fractions were obtained by elution using NACL salt gradient. All the main peak fractions were pooled together and concentrated by ultra filtration followed by dialysis. The purified protein was crystallized using hanging drop method. Rigaku R-AXIS IIc was used to measure the diffraction data of the triclinic crystal. Molecular replacement and crystallographic refinement of the pHSA triclinic crystal was implemented in X-PLOR. The crystallographic R factor of the current atomic model is 21.8 and 28.2% respectively. The atomic co-ordinates and structure factors of triclinic pHSA and rHSA have been deposited in the Brookhaven protein data bank with accession codes 1AO6 and 1BM0.
Results
Structural determination and quality
The structure of HSA was determined using to two different crystal forms with complementary properties. For solving the structure of protein, isomorphous replacement techniques were employed. pHSA consists of two HSA molecules each containing 5-582 residues and 7 water molecules whereas rHSA contains 5-282 residues and four water molecules. Due to conformational flexibility the electron density of residues 1-4 and 583-585 is not clearly observed. The Ramachandran plot reveals that all the residues fall into either the energetically favorable and allowed regions. The average temperature factors differ from residue to residue,the exposed ones had temperature factors of 100 Å2, while that for buried ones is around 15-20Å2.The overall temperature factors are 48.5Å2 for pHSA and 40.7Å2 for rHSA.
Local symmetry in the triclinic lattice
The asymmetric unit of the triclinic crystal consists of two HSA molecules. The local twofold symmetry is strict with a rotational angle of 179.8 degrees and translation vector length of less than 0.15 A for both pHSA and rHSA. The two molecules have identical structures with r.m.s deviations at c alpha, backbone atoms and the hydrogen atoms of 0.28,0.31 and 0.66. The r.m.s. deviations for cα atoms is fairy constant throughout the polypeptide chain.
Overall structure
The structures of pHSA and rHSA are identical with an r.m.s deviation of 0.24A° at the c alpha atoms. HSA is a heart shaped protein with dimentions of 80 x 80 x 30 A° consisting of three domains I(1-195), II(196-383), and III(384-585. Each domain is further divided into two sub-domains a and b. The shapes of domain II and III are similar. Domains I and II are perpendicular to each other hence forming a T shape and domain III forms a Y shape with the domains I and II. Due to this arrangement the protein has a heart shape. The two C-terminus helices of domain III do not retain their conformation during complex formation as they move towards the outside of the molecule. The HSA molecule shows molecular flexibility in different conditions due to relative motion of its domains.
Subdomains
Subdomains a and b are composed of 6 and 4 α helices. The helix clusters present in both sub domains are similar in three dimensional structure and related by a pseudo two fold axis. The total number of helices in HAS molecule is 28. In HSA, eight double disulfide bridges are formed by the seventeen disulfide bridges by the 34 cysteine residues. The first bridge connects the h1 and h3 helices perpendicularly while h3 and h4 alligned parallel. Hence h2, h3 and h4 joins to form a curved wall at one side of the domain. The residues having aliphatic and aromatic side chains are grouped at the inner surface of the helix to form the hydrophobic core of the sub domain which is the toplogical feature seen in all subdomains except in Ia h3.
Free sulfhydryl group at cys34
In a loop between the helices Ia-h2 and Ia-h3, there is Cys 34 the only cysteine residue which does not participate in any disulfide bridges. In the blood stream the sulfhydryl group of Cys 34 is oxidized by cysteine or glutathione in 30-40% of HSA molecules to from an intermolecular disulfide linkage. Even the Cys 34 located at the surface of the protein its Sγ atom is towards the interior and surrounded by side chains of Pro35, His39, Val77, Tyr84 which prevents the sulfhydryl group from coupling with the external counterparts. Ion spray mass spectroscopy reveals that pure HSA crystals used for crystallization contains few oxidized species at Cys 34 allows the growth of triclinic crystals. The backbone conformation may change to bring the free sulfhydryl group toward the exterior of the protein.
Binding sites for drugs and other compounds
A part of the hydrophobic core is surrounded by the first few residues of the extended loop together with helices a-h5 and a-h6 to form a pocket in subdomain IIa and IIIa. Site I is said to be the binding site for salicylates, sulfonamides and many other drugs of subdomain IIa. The inside wall of the pocket is formed by the hydrophobic side chains and the entrance is surrounded by the positively charged residues Arg257, Arg222, Lys199, His242, Arg218, Lys195. Site II, putative binding site for tryptophane, thyroxine, octanoate and other drugs of subdomain IIIa. The backbone nitrogens of the first three residues as well as the imidazole of His3 forms a binding site of the Cu+2. HSA is able to accommodate fatty acids. Arg 117 is located in a crevice formed by the helix cluster of subdomain Ib and the extended loop between Ia and Ib.
Conclusion
The X-ray structures of HSA derived from human pooled plasma or from a Pichia pastoris expression system, at a resolution of 2.5 Å, their three dimensional structures are identical within the margin of experimental error. HSA consists of three similar helical domains with eight pairs of double disulfide bridges. Deep hydrophobic pockets with positively charged entrances are located at similar positions in subdomains IIa and IIIa. There are three possible binding sites for long-chain fatty acids located on the surface of the molecule but their structural environments are entirely different.
3D structures of albumin
References
1) Protein Engineering, Vol. 12, No. 6, 439-446, June 1999�© 1999 Oxford University Press � Crystal structure of human serum albumin at 2.5 Å resolution S. Sugio1, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi
2) R. Beauchemin,1 C. N. N’soukpoé-Kossi,1 T. J. Thomas,2 T. Thomas,2 R. Carpentier,1,2 and H. A. Tajmir-Riahi1* Polyamine Analogues Bind Human Serum Albumin Biomacromolecules. 2007 October; 8(10): 3177–3183
3) JBiomol Struct Dyn. 2006 Dec;24(3):277-83.Resveratrol binding to human serum albumin. N' soukpoe-Kossi CN, St-Louis C, Beauregard M, Subirade M, Carpentier R, Hotchandani S, Tajmir-Riahi HA.
4) Biol Chem. 2000 Dec 8;275(49):38731-8. Binding of the general anesthetics propofol and halothane to human serum albumin. High resolution crystal structures. Bhattacharya AA, Curry S, Franks NP.
5) PDB files for HSA.
Proteopedia Page Contributors and Editors (what is this?)
Bhagiradhi Somalanka, Irene Becerra, Neeharika Pothuri, Michal Harel, Valerie Orovwigho