We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
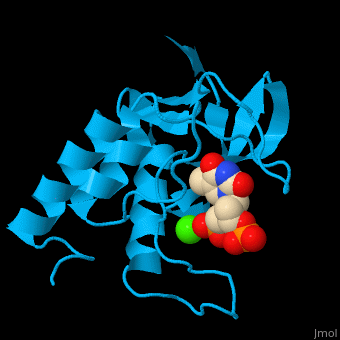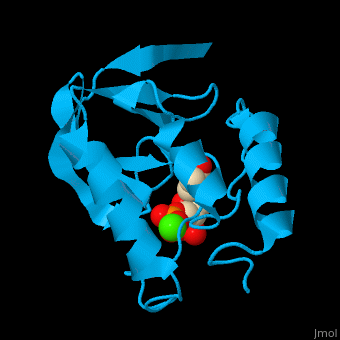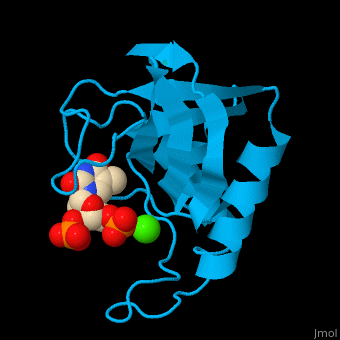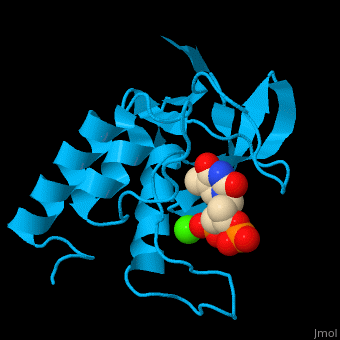
Staphylococcal nuclease
From Proteopedia
| |||||||||||
References
- ↑ Arnone AA, Bier CJ, Cotton FA, Day VW, Hazen EE Jr, Richardson DC, Richardson JS, Yonath A (1971) "A High Resolution Structure of an Inhibitor Complex of the Extracellular Nuclease of Staphylococcus aureus: I. Experimental Procedures and Chain Tracing," J. Biol. Chem. 246, 2303-2316
- ↑ Roche J, Caro JA, Norberto DR, Barthe P, Roumestand C, Schlessman JL, Garcia AE, Garcia-Moreno E B, Royer CA. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci U S A. 2012 Apr 10. PMID:22496593 doi:10.1073/pnas.1200915109
- ↑ Chen J, Lu Z, Sakon J, Stites WE. Proteins with simplified hydrophobic cores compared to other packing mutants. Biophys Chem. 2004 Aug 1;110(3):239-48. PMID:15228960 doi:10.1016/j.bpc.2004.02.007
- ↑ Wynn R, Harkins PC, Richards FM, Fox RO. Mobile unnatural amino acid side chains in the core of staphylococcal nuclease. Protein Sci. 1996 Jun;5(6):1026-31. PMID:8762134 doi:http://dx.doi.org/10.1002/pro.5560050605
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky, Jane S. Richardson