ToxT
From Proteopedia
IntroductionToxT or toxin co-regulated pilus virulence regulatory protein is a molecule at the end of a transcriptional cascade that autoregulates the transcription of the primary virulence factors of Vibrio cholerae[1] and itself. ToxT is a cytoplasmic protein that is activated in turn by ToxR, which is itself activated by ToxS in response to environmental stimuli.[2] These two factors, cholera toxin (CT)[2] and the toxin co-regulated pilus (TCP), are instrumental in causing the disease cholera[3]. This is an intestinal infection resulting in massive water loss in the affected individual, causing extreme dehydration.[4] Rehydration is sufficient as treatment. Structural FeaturesDNA-binding. ToxT belongs to a family of transcriptional regulators headed by and known as AraC.[1] The AraC family is characterized by a 100 amino acid region of sequence similarity that forms a with two helix-turn-helix motifs (one on either side of the black linker). [3] This DNA binding domain is composed of seven alpha helices. HTH1 is composed of alpha helices five and six, while HTH2 is composed of alpha helices eight and nine. The two HTH regions are linked by the very polar alpha helix seven(shown in black). The overall domain is located at the C-terminus.[1] Assuming ToxT is similar in mechanism to other AraC proteins, helix six from HTH1 and helix nine from HTH2 become aligned with the help of helix seven. Helix seven is positioned to attach to the N terminal binding pocket(the polar linking region) to allow binding to major consecutive grooves of target DNA (specific promoters for virulence genes).[1][5]. The conformation of helix seven is dependent on the ligand bound.
 MSA 1xtc
LigandIn this resolved structure, [9] is shown, which can be bound in the beta sheet barrel (as discussed above). This unsaturated fatty acid reduces virulence expression in Vibrio cholerae. Further StudyConclusive results about what activates ToxT itself has not yet been found. The varying activity of ToxT dependent on the presence of cis-palmitoleate or other unsaturated fatty acids represents a detailed method of effective pathogenicity in humans, but may not be a reasonable target for drug treatment. By restricting transcription (and thus translation and protein production) of virulence genes until the bacterium is determined to be in a favorable location for infection, Vibrio cholerae avoids wasting energy producing virulence factors that will just be cleared by the intestine. This is a specific mechanism to ensure that the bacterium also injects CT and TCP where they will do the most damage, perpetuating the infection. [6] Despite the lack of information about what activates ToxT itself, it is understood that the transcription of ctxA and tcpA by vibrio cholerae is sharply reduced in the presence of oleic, linoleic acid, and arachidonic acid, all of which are components of bile. Therefore, one may hypothesize that it may be possible to use the structure of a UFA or SFA to design a small molecule inhibitor of ToxT which may be used to treat or prevent cholera. EvolutionVibrio cholerae is a highly diverse species in which some strains are completely harmless, whereas other strains have the capacity to cause global cholera pandemics. It has been discovered that in each epidemic and pandemic strain, there is a chromosomal pathogenicity island (PAI) that is not present in the nonpathogenic strains. The region containing two ToxR-regulated genes (aldA and tagA) is composed of 13kb of previously unidentified DNA. [7]This region is part of a PAI that contains ToxT and a gene cluster a critical colonization factor and TCP. The PAI is 39.5 kb long, contains putative integrase and transposase genes, and inserts near a 10Sa RNA gene. One may infer that the PAI could have originated from a bacteriophage. This PAI was also found in two non-O1/non-O139 (which are both pandemic) sero type strains. Therefore, one may hypothesize that the PAI could be transferred within other strains of Vibrio cholerae. 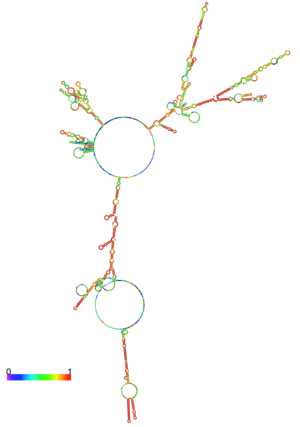 Centroid RNA 1xtc RNA StructureHere we have the centroid structure of the mRNA of ToxT. This mRNA is shown in its most stable conformation, with the less stable, higher energy regions in red. The lighter colored regions are more stable and lower in energy. 3D structure of ToxTUpdated on 20-August-2024 3gbg, 4mlo - VcToxT - Vibrio cholerae 6p7t - VcToxT (mutant) ReferencesBentivoglio, M; Pacini, P (1995). "Filippo Pacini: A determined observer". Brain Research Bulletin 38 (2): 161–5. doi:10.1016/0361-9230(95)00083-Q. PMID 7583342 Chiorazzo, G; Michael. Kull F; John. Lowden J; Michael. Pellegrini, Maria. Ronald, K; Taylor. Skorupski, Karen. (2009).“Structure of Vibrio Cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes.” 10.1073/pnas.0915021107 DiRita VJ, Parsot C, Jander G, Mekalanos JJ (June 1991). "Regulatory cascade controls virulence in Vibrio cholerae". Proc. Natl. Acad. Sci. U.S.A. 88 (12): 5403–7. doi:10.1073/pnas.88.12.5403. PMC 51881. PMID 2052618. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=51881. Sack DA, Sack RB, Nair GB, Siddique AK (January 2004). "Cholera". Lancet 363 (9404): 223–33. doi:10.1016/S0140-6736(03)15328-7. PMID 14738797. Klose, E; Karl. Osorio, R; Carlos. Prouty, G; Michael. (2005) “Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT” DOI: 10.1111/j.1365-2958.2005.04897.x Prevention and control of cholera outbreaks: WHO policy and recommendations, World Health Organization, Regional Office for the Eastern Mediterranean, undated but citing sources from ’07, ’04, ’03, ’04, and ’05. Bailey, C; Camilla. Boedeker, C; Edgar. Johnson, A; Judith. Kaper, B; James. Karaolis, R; David. Reeves, R; Peter. (1997). “A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains.” March 17, 1998 vol. 95 no. 6 3134-3139 | ||||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Ingrid Youngworth, Yang Yang, Michal Harel, Alexander Berchansky, Jaime Prilusky