User:Abbas Raza/Sandbox 1
From Proteopedia
Contents |
NEIL3: a DNA repair glycosylase from Mus musculus
|
This protein is a unique member of DNA glysosylases, family of base excision repair enzymes that show preference for both oxidative purines and pyrimidine lesions. For a description of all known DNA glycosylases and their families see the wikepedia page on DNA glycosylases [[1]]. There is also a separate proteopedia page on Nei/fpg family [[2]].
Originally identified through in-silico studies from Wallace, Mitra and Seeger group back in 2002[1],[2],[3] the crystallisation of NEIL3 remained challenge for almost a decade due to the difficulty in getting a stabilized form of the protein for crystallization until a truncated version was crystallized in 2013 [4] and the crystal structure publlished as .
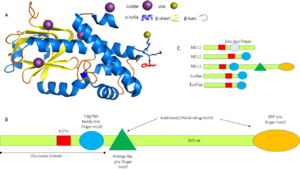
NEIL3 proteins are almost twice the size of other Fpg/Nei family members (Figure 1C). This protein belongs to α/β[[3]] category of proteins with the following secondary structure elements: , , , , , ,,,,, , ,,,, . The beta strands form a two-layered at the N-terminus in which each layer is composed of four anti-parallel beta strands. The C terminus has a Helix 2 turn Helix (H2TH) motif formed by and a canonical formed by the interconnecting loop between β strands-9 and 10. In contrast to other Fpg/nei members, NEIL3 has two additional DNA binding domains at their C-terminus: one Ran binding protein (RanBP2)-type zinc finger motif and two identical GRF-zinc finger motif but they were not available in the truncated 3w0f model. The catalytic residue of MmuNEILL3 is a rather than a proline as for other Fpg/nei members but serves the same purpose in catalysis and is surrounded by that forms the DNA binding cleft and lesion binding pocket of NEIL3.
3D structures of other Fpg/nei members
Updated on 28-April-2014
NEIL1
1tdh, 3a42, 3a45, 3a46, 3vk7, 3vk8, 4nrv, 4nrw
NEIL2
Fpg
3twk, 3twl, 3twm, 1pjj, 1r2y, 1ee8, 1k82, 1l1z, 1l2b, 1l2c, 1l1t, 1l2d, 1r2z, 1pm5, 1nnj, 1kfv, 1pji, 3c58, 3vk5, 2xzu, 2xzf, 1tdz, 1xc8
Nei
2eao, 2opf, 2oq4, 1q3c, 1q3b, 1q39
Biological Function
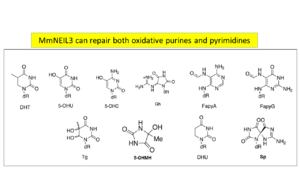
MmuNeil3 is a bifunctional DNA glycosylase that exhibits a broad substrate recognition spectrum (Sp, Gh, Fa pyG, FapyA, MeFapyG, DHU, DHT, 5-OHU, 5-OHC,5-OHMH, Tg, 8-oxoA, AP) but not 8-oxo-G (Figure2). . The reason being that α-B in Neil3 does not align with the corresponding helix in the other Nei families (Figure 2). Additional stabilizing bonds by residues Asp-155, Ile-156, and Ser-158 and neighboring residues Gly-81 and Arg-227 create a restricted movement and hence catalysis of predominately single stranded lesions by MmNEIL3 (Figure3)The DNA structures attacked most often are large bubble and single stranded DNA and least duplex DNA[5]. NEIL2 is similar to NEIL3 in terms of substrates attacked whereas NEIL1 on the other hand, prefers double stranded DNA lesions. The idea of having multiple enzymes to repair the same lesion confirms that NEIL3 might act as a backup line of defense against oxidative damage.
NEIL3 expression and localisation
The expression of NEIL3 is tissue specific with high levels in spleen, testis, bone marrow and brain. Transfection of Hela cell lines localised this protein to nucleus but not in mitochondria[6]. The protein levels fluctuate during cell cycle like other DNA glycosylases with highest expression during G2 phase of cell cycle[7].
Disease
Experiments with knock-out NEIL3-/-mice showed that the animals were viable and fertile and had no obvious contrasting phenotype[8] . However, later studies have reported memory defects and reduced anxiety like symptoms in C57BL/6 mice indicative of impaired proliferatived neural cells but no defects in fertility or viability[9]. This adds to the idea that the biological function of NEIL3 is not exclusively restricted to DNA repair. High expression in spleen and bone marrow hints a role in immune system.
References
- ↑ Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst). 2002 Jul 17;1(7):517-29. PMID:12509226
- ↑ Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3523-8. PMID:11904416 doi:http://dx.doi.org/10.1073/pnas.062053799
- ↑ Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002 Nov 15;30(22):4926-36. PMID:12433996
- ↑ Liu M, Imamura K, Averill AM, Wallace SS, Doublie S. Structural Characterization of a Mouse Ortholog of Human NEIL3 with a Marked Preference for Single-Stranded DNA. Structure. 2013 Feb 5;21(2):247-56. doi: 10.1016/j.str.2012.12.008. Epub 2013 Jan, 9. PMID:23313161 doi:http://dx.doi.org/10.1016/j.str.2012.12.008
- ↑ Liu M, Doublie S, Wallace SS. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat Res. 2013 Mar-Apr;743-744:4-11. doi: 10.1016/j.mrfmmm.2012.12.003. Epub, 2012 Dec 26. PMID:23274422 doi:http://dx.doi.org/10.1016/j.mrfmmm.2012.12.003
- ↑ Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005 Dec;138(6):763-72. PMID:16428305 doi:http://dx.doi.org/10.1093/jb/mvi168
- ↑ Bar-Joseph Z, Siegfried Z, Brandeis M, Brors B, Lu Y, Eils R, Dynlacht BD, Simon I. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc Natl Acad Sci U S A. 2008 Jan 22;105(3):955-60. doi:, 10.1073/pnas.0704723105. Epub 2008 Jan 14. PMID:18195366 doi:http://dx.doi.org/10.1073/pnas.0704723105
- ↑ Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005 Dec;138(6):763-72. PMID:16428305 doi:http://dx.doi.org/10.1093/jb/mvi168
- ↑ Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjoras M. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Natl Acad Sci U S A. 2011 Nov 15;108(46):18802-7. doi:, 10.1073/pnas.1106880108. Epub 2011 Nov 7. PMID:22065741 doi:http://dx.doi.org/10.1073/pnas.1106880108