User:Alice Harmon/Sandbox 4
From Proteopedia
Contents |
SnRK2.6/OST1/SRK2E
Role in Abscisic Acid signaling
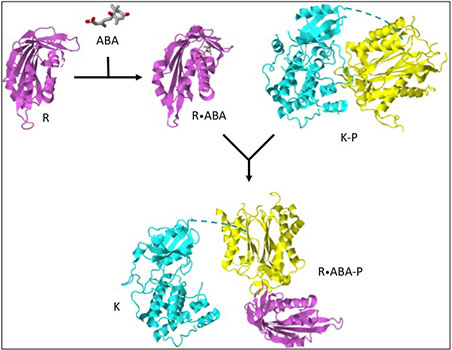
SnRK2.6/OST1/SRK2E is a triply-named protein kinase from Arabidopsis, which is activated by the abscisic acid (ABA) response pathway[1][2][3][4]. As shown in the figure, in unstimulated cells SnRK2.6/OST1/SRK2E (K) and a protein phosphatase 2C (P) are in a complex (K-P) in which the kinase is dephosphorylated and inactive[5][6]. The kinase and phosphatase are proposed to be tethered via the C-terminal sequence (dashed line) of the kinase[5][6][7]. When ABA levels in the cytosol rise, ABA binds to an ABA receptor (R)[8][9][10]. The activated receptor (R.ABA) binds to the protein phosphatase (R.ABA-P) and inactivates it. SnRK2.6/OST1/SRK2E, although still tethered to the phosphatase, is now free to be activated by autophosphorylation or phoshorylation by another protein kinase[11][12]. Activation of SnRK2.6/OST1/SRK2E leads to phosphorylation of: 1) ion channels SLAC1[13] and KAT1[14] in guard cells and stomatal closure; 2) transcription factor ABI5[3] in seeds/seedlings and dormancy/growth arrest; or 3) phosphorylation of transcription factor AREB/ABF [15][4] in vegetative tissue and stress tolerance and growth regulation.
Structures in the figure are: R, apo pYR2, 3kdh; R.ABA, PYR2.ABA, 3kdi; K-P; SnRK2.6-HAB1, 3ujg; R.ABA-P, PYR2.ABA-ABI2, 3ujl; K, SnRK2.6,3uc4.
Kinase names and family members
Two of SnRK2.6/OST1/SRK2E's three names originated from its membership in subclass III of the SnRK2 family of protein kinases. It was named SnRK2.6 by Hrabak et al.[16] and SRK2E by Umezawa et al.[5]. SnRK2 stands for SNF1-related kinase group 2, which in Arabidopsis has 10 members. SNRK2s are members of the calmodulin-dependent protein kinase clade of protein kinases. The third name OST1 (open stomata 1)[1] is descriptive of the phenotype of plants bearing a gene mutation that produces an inactive protein kinase.
Two other family members in Arabidopsis, SNRK2.2/SRK2D and SnRK2.3/SRK2I, are activated by the ABA pathway in the same manner as SnRK2.6. Each of these kinases interacts with a member of clade A of the protein phosphatase 2C family - ABI1, HAB1 or HAB2. In rice homologs of these protein kinases are named SAPK8, SAPK9 and SAPK10.
Kinase structure and regulation
SnRK2.6/OST1/SRK2E has a primary structure comprising an amino terminal Eukaryotic Protein Kinase Catalytic Domain and a C-terminal sequence that contains the SNRK2 box, which is unique to the SNRK2 family and required for activity[17][18]. Its C-terminus also contains a sequence called the ABA box, which is found only in the family members that are responsive to ABA[18]
| Left scene - unphosphorylated SnRK2.6 without any ligands 3uc4[17] | Right scene - SnRK2.6 (blue) in complex with the protein phosphatase 2C, HAB1 (gold), with Mg2+ and SO42- 3ujg[6] | ||||||||||||
3uc4 scenes The catalytic domain of SnRK2.6 is typical of Eukaryotic Protein Kinase Catalytic Domain) except for an additional α-helix (shown as strands) in the small lobe, which is formed by SNRK2 box sequence. The activation segment (with unresolved gap), including the D of the DFG motif in ball and stick, is blue. The catalytic loop, including the D of the DLKLEN motif in ball and stick, is orchid. Subdomain III, including its invariant E in ball and stick, is gold. The invariant K of subdomain II is in chartreuse. The SnRK2 box is turquoise. The C-terminal domain, that includes the ABA box is unresolved. The arrangement of the residues in ball and stick around the active site, indicate that this structure is in a partially active state in spite of its unphosphorylated activation loop. This is possibly due to the interaction of the SNRK2 box helix with subdomain III.[17]. The interaction between these helices is similar to the interaction of helices in the complex between 1w98. Here we see that subdomain III of the protein kinase (opaque blue) is stablized by interaction with a helix from cyclin (opaque gold). The positioning of subdomain III by this interaction is critical for for formation of the active site.[19]. |
3ujg scenes The two enzymes are bound via interface their active sites. The phosphatase inactivates the kinase by dephosphorylating the kinase activation loop and by sterically blocking the kinase active site. The complex was constructed as a fusion protein with a 6His-tag at the N-terminus of SnRK2.6 (residues 11–362) fused to HAB1(172–511) via a GSGSAGSAAGS linker. Mutations of D296A and E297A in SnRK2.6 were introduced at the crystal packing interface to reduce surface entropy. The same structures as in the left scene are shown. The fully resolved activation segment extends into the phosphatase's active site and is unphosphorylated. Residues 319-362 of SnRK2.6, which includes the ABA box, and the GSGSAGSAAGS linker are not resolved. The disorganization of the residues shown in ball and stick, with most pointing away from the active site, indicates that the catalytic domain is in the inactive state. The activation loop (blue trace) of SnRK2.6 is inserted into the catalytic site (marked by the magnesium ions) of the phosphatase. The phosphorylatable residue of the activation loop S175 (CPK ball and stick) is positioned near the magnesium ions. W385 of the phosphatase (brown ball and stick) in turn protrudes into the kinase's active site, where it interacts with residues R139 and Glu144 (CPK ball and stick) of the catalytic loop (orchid trace) and I183 of the activation loop. SnRK2.6 is shown in blue cartoon, and HAB1 in gold spacefill. The ABA box sequence is not resolved, but it would extend from the C-terminal end of the SNRK2 box helix (cyan helix). It is proposed that the ABA box sequence, which is highly acidic, binds to a patch of basic residues (blue) on the surface of the phosphatase.[6] |
SNRK2 structures
3uc3 Arabidopsis thaliana SNRK2.3 + Co2+
3zut AtSNRK2.6 (D160A mutant)+ ANP
3zuu AtSNRK2.6 (D160A, S175D mutant) + gold
3uc4 apoAtSNRK2.6 (D59A, E60A mutant)
3udb apoAtSNRK2.6 (C131A, C157A, C159A, S7A, s29A, s43A, S166A, T175A)
complex with a protein phosphatase 2C
3ujg AtSNRK2.6 (D296A) + HAB1 + Mg2+
References
- ↑ 1.0 1.1 Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002 Dec;14(12):3089-99. PMID:12468729
- ↑ Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002 Dec;43(12):1473-83. PMID:12514244
- ↑ 3.0 3.1 Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009 Jul;50(7):1345-63. doi: 10.1093/pcp/pcp083. Epub 2009, Jun 18. PMID:19541597 doi:10.1093/pcp/pcp083
- ↑ 4.0 4.1 Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007 Feb;19(2):485-94. Epub 2007 Feb 16. PMID:17307925 doi:tpc.106.048538
- ↑ 5.0 5.1 5.2 Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009 Oct 13;106(41):17588-93. doi:, 10.1073/pnas.0907095106. Epub 2009 Sep 29. PMID:19805022 doi:10.1073/pnas.0907095106
- ↑ 6.0 6.1 6.2 6.3 Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, Brunzelle JS, Zhang H, Yang H, Jiang H, Li J, Yong EL, Cutler S, Zhu JK, Griffin PR, Melcher K, Xu HE. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012 Jan 6;335(6064):85-8. Epub 2011 Nov 24. PMID:22116026 doi:10.1126/science.1215106
- ↑ Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006 Feb 24;281(8):5310-8. Epub 2005 Dec 19. PMID:16365038 doi:M509820200
- ↑ Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009 Dec;16(12):1230-6. Epub 2009 Nov 5. PMID:19893533 doi:10.1038/nsmb.1730
- ↑ Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009 May 22;324(5930):1064-8. doi: 10.1126/science.1172408. Epub 2009, Apr 30. PMID:19407143 doi:10.1126/science.1172408
- ↑ Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009 May 22;324(5930):1068-71. doi: 10.1126/science.1173041. Epub 2009, Apr 30. PMID:19407142 doi:10.1126/science.1173041
- ↑ Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol. 2007 Mar;63(4):491-503. PMID:17103012 doi:10.1007/s11103-006-9103-1
- ↑ Burza AM, Pekala I, Sikora J, Siedlecki P, Malagocki P, Bucholc M, Koper L, Zielenkiewicz P, Dadlez M, Dobrowolska G. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J Biol Chem. 2006 Nov 10;281(45):34299-311. Epub 2006 Sep 15. PMID:16980311 doi:10.1074/jbc.M601977200
- ↑ Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, Romeis T, Hedrich R. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci U S A. 2009 Dec 15;106(50):21425-30. doi:, 10.1073/pnas.0912021106. Epub 2009 Dec 2. PMID:19955405 doi:10.1073/pnas.0912021106
- ↑ Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, Uozumi N. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009 Dec 10;424(3):439-48. doi: 10.1042/BJ20091221. PMID:19785574 doi:10.1042/BJ20091221
- ↑ Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci U S A. 2006 Feb 7;103(6):1988-93. Epub 2006 Jan 30. PMID:16446457 doi:10.1073/pnas.0505667103
- ↑ Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003 Jun;132(2):666-80. PMID:12805596 doi:10.1104/pp.102.011999
- ↑ 17.0 17.1 17.2 Ng LM, Soon FF, Zhou XE, West GM, Kovach A, Suino-Powell KM, Chalmers MJ, Li J, Yong EL, Zhu JK, Griffin PR, Melcher K, Xu HE. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21259-64. Epub 2011 Dec 12. PMID:22160701 doi:10.1073/pnas.1118651109
- ↑ 18.0 18.1 Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006 Aug;141(4):1316-27. Epub 2006 Jun 9. PMID:16766677 doi:pp.106.079327
- ↑ Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, Brown NR, Johnson LN. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005 Feb 9;24(3):452-63. Epub 2005 Jan 20. PMID:15660127
See Also
[1] Abscisic Acid in Wikipedia