User:Cameron Ball/Sandbox 1
From Proteopedia
Replication Terminator Protein (RTP) is a protein found in Bacillus Subtilis(B.Subtilis) that plays an important role in the termination of bacterial chromosome replication. RTP binds to the circular bacterial genome to block the progression of DNA polymerase in a polar manner. A protein that performs the same function as RTP has been found in Eschericia coli (E.coli), named “Termination Utilisation Substance” (Tus)
Contents |
Introduction
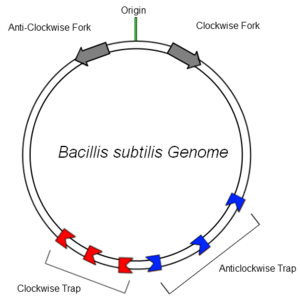
The bacterial genome of B.subtilis is circular and contains only one origin of replication (OriC). In order to increase the efficiency of DNA replication, the DNA is copied in clockwise and anticlockwise directions simultaneously and later ligated together. RTP bound to Ter sites in the clockwise and anticlockwise traps arrests the respective replication fork and aids termination of replication, preventing continuous copying of the genome. It has been found that many organisms employ a mechanism to aid in this termination, suggesting an evolutionary advantage in possessing such a system [1](although mutants lacking the tus gene are still viable[2]).
The Termination Sites
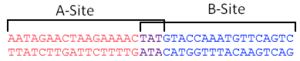
To arrest DNA replication, the two RTP dimers bind at a specific DNA site. These sites, christened Ter sites, are 29 base pairs in length and comprise two non-identical inverted repeats that share three highly conserved base pairs. The two sites are designated the A-site and the B-site and each have different affinities for RTP (with the B-site exhibiting greater protein-DNA contact and thus greater affinity[3]). Each site binds one dimer of RTP cooperatively, with the higher affinity B-site being bound first, then the A-site. [4]
It is now known that RTP binds to Ter site in a directional manner as a result of these non-identical sites, which plays an important role in allowing polymerase units to pass in one direction, but blocking them in another. However, historically, the exact way in which symmetric RTP dimers blocked polymerases in a polar fashion was a source of great confusion. This was compounded by the fact that a symmetrical DNA sequence (designated sRB) was used in the first crystal structure of an RTP:DNA complex. [5]
B.subtilis has multiple Ter sites of both polarities to ensure that replication is terminated. The sites are situated on either side of the diff site (opposite the OriC) to provide some redundancy at the recombination site. This ensures the entire genome is copied faithfully.
Structure
|
through interactions between the and the major groove of DNA. The first crystal structure of RTP complexed to the native TerB site (nRB) was solved by vivian et al in 2007[6]. The induced asymmetry of the RTP dimer results in a and a beta loop turn. These loops are thought to be important in the cooperative binding of two RTP dimers, by physically contacting the DNA molecule and promoting DNA bending.[7][8]
Mechanism
Two possible (non-mutually exclusive) mechanisms were originally proposed to explain it's activity; The "differential binding affinity" model (DBA), where RTP's different affinity for the A and B sites gives rise to the polarity, and the "conformational change" model, where the polarity is a result of different conformations assumed by each RTP dimer on each half site.[9] Recent data has shown that RTP-Ter contact is not sufficient to cause arrest, suggesting there is more to each proposed model[10][11]. Additionally, in 2006 Ian Duggin showed that the fork arrest efficiency of RTP can be lowered while maintaining the same RTP-DNA binding affinity by attaching short peptides to RTP's non-permissive face. These peptides block any protein-protein interaction with the approaching replisome unit [12] suggesting that RTP is not simply acting as a directional block, but that there is some protein-protein interaction occuring that has yet to be elucidated.
To summarise, one RTP dimer binds at the B-site and deforms the DNA in such a way that binding of another dimer at the A site is favourable, and the different affinities of the two sites for RTP results in non symmetrical binding. When a polymerase unit approaches the RTP from the permissive face (A site) the RTP complex is dislodged and replication continues. If the polymerase approached from the non permissive face the stronger interaction of the B-site with RTP, in combination with a protein-protein interaction (possibly with the approaching helicase, halting DNA melting) arrests DNA replication.
Comparison of Tus and RTP
As previously mentioned, E.coli express a protein called tus that performs an almost identical function to RTP. Despite the similarity of their purpose, the underlying mechanism of action and 3D structure are markedly different. This suggests that these two systems evolved separately to each other, once again highlighting their importance [13]. The differences between the two proteins are summarised below.
| RTP | Tus | |
|---|---|---|
| Structure | Binds as a symmetrical dimer | Binds as an asymetrical monomer |
| DNA Binding Mechanism | Contacts DNA major groove via alpha helices, bends DNA | Basic Cleft in Tus contacts major groove of DNA [14] |
| Mechanism of Action | Binds to Ter site directionally, bends DNA and may contact DnaB protein directly to inhibit DNA melting | Contacts DnaB to halt it's translocation along DNA [15] |
| Ter Site | 29 bp site, contains two 16bp non-identical inverted repeats (A and B sites) that share a 3 base pair overlap, each site binds a dimer of RTP | 23 bp site, no internal symmetry, binds a monomer of tus. |
In 2000, Anderson et al inserted the E.coli TerB sequence into a B.subtilis plasmid and transformed it into a B.subtilis cell line that could overexpress tus. They found that the Tus-Ter system arrested fork activity in B.subtilis with approximately 5% efficiency, compared to 45% efficiency in E.coli. They also performed the reverse experiment, by introducing the B.subtilis RTP-Ter system into E.coli. They found that fork arrest efficiency was approximately 3.2%, about a 3 fold decrease over the native activity in B.subtilis (10.6%) [16]
This once again emphasises the difference of the two systems and also confirms that simple RTP/Tus-Ter contact is not enough to efficiently cause fork arrest.
References
- ↑ A.A. Griffiths, P.A. Andersen and R.G. Wake, Replication terminator protein-based replication fork-arrest systems in various Bacillus species, J. Bacteriol. 180 (1998), pp. 3360–3367
- ↑ King, G.F., Wake, R.G., A tale of two terminators: crystal structures sharpen the debate on DNA replication fork arrest mechanisms 1997 Structure Vol 5 No 1
- ↑ Langley, D. B., Smith, M. T., Lewis, P. J., and Wake, R. G. Protein-nucleoside contacts in the interaction between the replication terminator protein of Bacillus subtilis and the DNA terminator. (1993) Mol. Microbiol. 10, 771-779
- ↑ A.V. Kralicek, P.K. Wilson, G.B. Ralston, R.G. Wake and G.F. King, Reorganization of terminator DNA upon binding replication terminator protein: implications for the functional replication fork arrest complex, Nucl. Acids Res. 25 (1997), pp. 590–596
- ↑ Wilce, J. A., Vivian, J. P., Hastings, A. F., Otting, G., Folmer, R. H., Duggin, I. G., Wake, R. G. & Wilce, M. C., Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest., (2001). Nature Struct. Biol.8, 206–210.
- ↑ J.P. Vivian, C.J. Porter, J.A. Wilce, M.C.J. Wilce, An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA, Journal of Molecular Biology, Volume 370, Issue 3, 13 July 2007, Pages 481-491, ISSN 0022-2836, DOI: 10.1016/j.jmb.2007.02.067.
- ↑ J.P. Vivian, C.J. Porter, J.A. Wilce, M.C.J. Wilce, An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA, Journal of Molecular Biology, Volume 370, Issue 3, 13 July 2007, Pages 481-491, ISSN 0022-2836, DOI: 10.1016/j.jmb.2007.02.067.
- ↑ A.V. Kralicek, P.K. Wilson, G.B. Ralston, R.G. Wake and G.F. King, Reorganization of terminator DNA upon binding replication terminator protein: implications for the functional replication fork arrest complex, Nucl. Acids Res. 25 (1997), pp. 590–596
- ↑ Duggin, I.G., Mathews, J.M., Dixon, N.E., Wake, R.G., Mackay, J.P., A Complex Mechanism Determines Polarity of DNA Replication Fork Arrest by the Replication Terminator Complex of Bacillus subtilis 2005 The Journal of Biological Chemistry, 280, 13105-13113.
- ↑ Duggin, I.G., Mathews, J.M., Dixon, N.E., Wake, R.G., Mackay, J.P., A Complex Mechanism Determines Polarity of DNA Replication Fork Arrest by the Replication Terminator Complex of Bacillus subtilis 2005 The Journal of Biological Chemistry, 280, 13105-13113.
- ↑ Kaplan, D.L and Bastia, D, Mechanisms of polar arrest of a replication fork, Molecular Microbiology 2009:72:2), pp 279-285
- ↑ Duggin, I.G., DNA replication fork arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding J Mol Biol 361:1-6
- ↑ King, G.F., Wake, R.G., A tale of two terminators: crystal structures sharpen the debate on DNA replication fork arrest mechanisms 1997 Structure Vol 5 No 1
- ↑ Kaplan, D.L and Bastia, D, Mechanisms of polar arrest of a replication fork, Molecular Microbiology 2009:72:2), pp 279-285
- ↑ Kaplan, D.L and Bastia, D, Mechanisms of polar arrest of a replication fork, Molecular Microbiology 2009:72:2), pp 279-285
- ↑ Andersen, P., Griffiths, A., Duggin, I., and Wake, R. (2000) Functional specificity of the replication fork-arrest complexes of Bacillus subtilis and Escherichia coli: significant specificity for Tus-Ter functioning in E. coli. Mol Microbiol 36: 1327–1335.