HIV-Protease
Human Immunodeficiency Virus (HIV) is the cause of Acquired Immunodeficiency Syndrome (AIDS). HIV directs the synthesis of several polyproteins, which each consist of several tandemly linked proteins. The maturation of the virus to its infectious form requires that these polyproteins be cleaved to their component proteins. , a homodimeric enzyme, is responsible for doing so and is therefore crucial to the virus's infectious capacity.
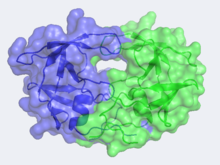
Structure of HIV-1 Protease
The X-ray structure of HIV-1 protease reveals that it is composed of , each consisting of 99 amino acid residues. The subunits come together in such as way as to . This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of , making it a member of the aspartyl protease family. The two Asp's are that activate a water molecule to hydrolytically cleave the polyprotein that binds in the tunnel.[1] You may be wondering how a polyprotein makes its way into the active-site tunnel, as the to admit it. The key is the two flexible flaps on the top of the tunnel that to enter the tunnel. The flaps , shifting from an open to a closed conformation to bind the target in an appropriate conformation for cleavage.
Medical Implications
There currently is no cure or vaccine against HIV. Researchers, however, have discovered treatments that can halt and even reverse the progression of AIDS, due in large part to our understanding of the structure of HIV-1 protease. was the first protease inhibitor approved by the FDA for the treatment of HIV. It inhibits HIV protease by , preventing the binding of polyproteins. Its chemical structure mimics the tetrahedral intermediate of the hydrolytic reaction, thereby .[2] Saquinavir is essentially an uncleavable ligand, as indicated by the on binding saquinavir or a polypeptide . Other drugs used to treat HIV infection that inhibit include (1hsg), (1hxw), and (1ohr).