User:Eran Hodis/1ea5 test
From Proteopedia
Acetylcholinesterase (E.C. 3.1.1.7)
Contents |
Overview
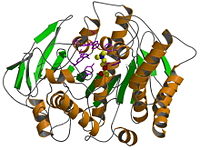
The 3D structure of acetylcholinesterase (AChE) from Torpedo californica electric organ has been determined by x-ray analysis to 1.8 Å resolution. The form crystallized is the glycolipid-anchored homodimer that was purified subsequent to solubilization with a bacterial phosphatidylinositol-specific phospholipase C. The enzyme monomer is an α/β structure protein that contains 537 amino acids. It consists of a 12-stranded mixed beta sheet surrounded by 14 α-helices and bears a striking resemblance to several hydrolase structures including dienelactone hydrolase, serine carboxypeptidase-II, three neutral lipases, and haloalkane dehalogenase. The is unusual because it contains Glu, not Asp, in the Ser-His-acid catalytic triad and because the relation of the triad to the rest of the protein approximates a mirror image of that seen in the serine proteases. Furthermore, the active site lies near the bottom of a deep and narrow gorge that reaches halfway into the protein. Modeling of acetylcholine binding to the enzyme suggests that the quaternary ammonium ion is bound not to a negatively charged "anionic" site, but rather to some of the 14 aromatic residues that line the gorge.
Biological context
AChE (E.C. 3.1.1.7) rapidly hydrolyzes acetylcholine released into the synapse. Its catalytic activity can be represented as acetylcholine + H2O = choline + acetate. AChE is found in the synapses and at neuromuscular junctions, and on erythrocyte membranes.
Additional information
1EA5 hydrolyzes choline released into the synapse. Catalytic activity: acetylcholine + H2O = choline + acetate. Inhibitors of the enzyme AChE slow and sometimes reverse the cognitive decline experienced by individuals with Alzheimer's disease. T californica AChE is a G2 dimer in solution (see Sussman 1988). The asymmetric unit contains a monomer, with the crystallographic 2-fold axis relating the two monomers in a dimer. This is the highest resolution AChE determined so far. There is recent evidence (see Bucht 1996) that the gpi anchor is attached to either ser 543 or ser 544, not to sys 537.
Related diseases
Blood group, Yt system, Endplate acetylcholinesterase deficiency
PDB entry information
Exp. Method: X-ray diffraction
Authors: Michal Harel, M.Weik, I.Silman, Joel L. Sussman
Date: Nov 8, 2000
Full crystallographic information is available from OCA.
Links
1ea5 is a Single protein structure of sequence from Torpedo californica. Active as Acetylcholinesterase, with EC number 3.1.1.7
References
- Bucht, G., Hjalmarsson, K., (1996) "Residues in Torpedo californica acetylecholinesterase necessary for proessing to a glycosyl phosphatidylinositol-anchored form", Biochim Biophys Acta 1292 (223).
- Axelsen, P.H., Harel, M., Silman, I., Sussman, J.L., (1994) "Structure and dynamics of the active site gorge of acetylcholinesterase: synergistic use of molecular dynamics simulation and X-ray crystallography", Protein Sci. 3 (188).