Introduction
is an understudied protein, and therefore very little is known about its overall function. It is a nuclear protein that is a highly sequentially and structurally conserved paralog to ATAD2 [1]. It is unknown when the gene duplication event occurred that was able to form both ATAD2 and ATAD2B proteins in evolutionary history, or how ATAD2 and ATAD2B interact with each other within a cellular context. ATAD2 is a nuclear co-regulator protein and found to be highly overexpressed in many unrelated forms of cancer[2][3]. ATAD2 overexpression is linked to poor prognosis in these cancer patients. In cancer cell proliferation, ATAD2 is a direct target of E2F, a member of the frequently mutated retinoblastoma protein pathway[4]. ATAD2 also acts as a cofactor for the Myc oncogene, and is engaged in a positive feedback loop to continually amplify the expression of itself and other coregulators in cancer[5]. The overall biological function of ATAD2 has yet to be elucidated, but has been implicated in stimulating gene expression, promoting chromatin remodeling, and impacting DNA repair. On the other hand, ATAD2B has not been characterized nearly as much as ATAD2, but it is thought to be involved in neuronal development, and was found overexpresed in multiple human tumors[6][7].
Structural Organization
Both the ATAD2 and ATAD2B proteins contain two conserved domains: two AAA ATPase domains (AAA1+AAA2) and a bromodomain[8]. The ATPase domains are associated with diverse cellular activities and are thought to play a role in ATAD2 oligomerization, and are suspected to act as a molecular motor involved in chromatin remodeling. Bromodomains "read" or interpret the epigenetic acetylated lysine post-translational modification (PTM)[9].
Investigators who study ATAD2 and ATAD2B focus on their bromodomains, because they can be targeted pharmaceutically for disease therapy. The structure and function of the ATAD2 bromodomain has been well characterized[10] . The binding function of the ATAD2B bromodomain has begun to be characterized[11].
While there are over 100 ATAD2 bromodomain structures in the PDB, and some other domains have been characterized in yeast homologs[12], the bromodomain is the only domain to be studied or characterized structurally, with only three structures in the PDB for ATAD2B.
Although ATAD2 and ATAD2B are highly conserved, there is very little known about the function of ATAD2B, the function of its domains, or its role in oncogenesis.
ATAD2B Function
Many players, little knowledge...
Role of histone post-translational modifications (PTMs)
Epigenetics is the study of the molecules or mechanisms that cause changes in gene expression, without any modification to the underlying DNA sequence. DNA methylation, long non-coding RNAs, and post-translational modifications (PTMs) are all areas of epigenetic study [13].
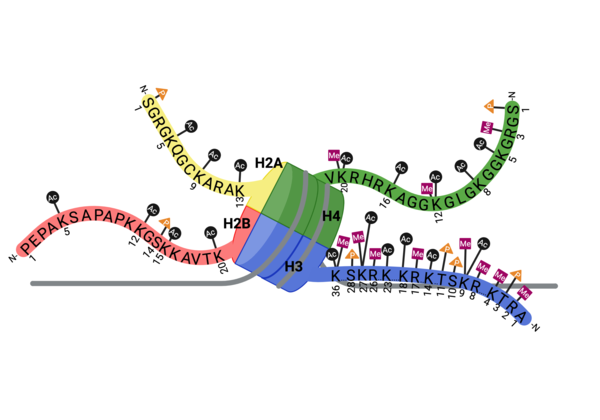
Visual illustrating the NCP compacting DNA into higher order chromatin structure.
PTMs can occur on the histone proteins that compose the (NCP). The NCP is the fundamental unit of chromatin and impacts gene accessibility to transcriptional regulators. Histones are positively charged and work to compact the negatively charged DNA around an octamer of histone proteins. Multiple NCPs compact DNA to form higher order chromatin structure, all the way to the level of the chromosome[14]. Additionally, there are unstructured extensions at the N-terminus of histone proteins, termed , which are also positively charged. This compact histone octamer wrapped with DNA becomes the nucleosome core particle (NCP), the fundamental unit of chromatin, which poses a barrier to transcription[15][16].
Multiple types, numbers, and combinations of PTMs can occur on the histone tails of the nucleosome at any given time. PTMs can serve as docking sites for protein effectors, which impact chromatin structure and gene expression[17][18]. Methylation, phosphorylation, and acetylation have been the most well characterized. Methylation modifications provide a platform for regulatory factors associated with gene activation or repression, depending on the modified residue. Phosphorylation ensures proper chromatin compactio during meiosis and mitosis, as well as defining the site of DNA damage. Acetylation neutralizes the positive charge on lysine and arginine residues, loosening histone:DNA packing, for active gene expression. These positively charged lysine residues are found on the tails of histone proteins, and once acetylated, the positive charge the lysine residue carries is removed, which can loosen the DNA compaction around the central octamer of histone proteins and expose a target gene on the DNA make it more amenable to transcription factors.
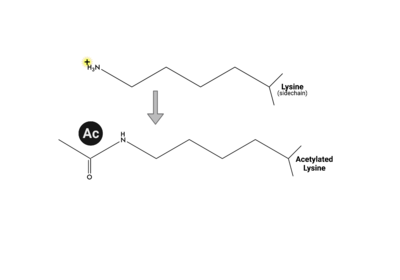 When lysine is acetylated, its positive charge becomes neutralized. | 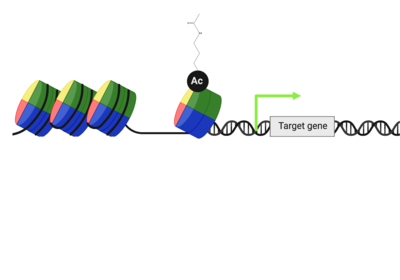 This lysine acetylation impacts the packing interactions between histones & DNA, making a target gene more accessible to transcription factors. |
The bromodomain function is conserved
The only known function of the bromodomain is to bind to acetylated lysine residues. This bromodomain function is highly conserved throughout evolution and across 42 bromodomain-containing proteins. Bromodomains can also serve as a site to recruit transcriptional machinery to the target gene once accessible, thus having the power to impact gene expression. Bromodomains have been known about since the 1990s, and in 2012, Filippakopoulos, et al. characterized and divided these bromodomains into eight sub-families based upon similarities in sequence and structure [19]. ATAD2 and ATAD2B are members of subfamily IV, along with other bromodomain-containing proteins, the BRPF(BRomodomain and PHD Finger containing proteins) family. Despite bromodomains having the same distinct structural fold, the sequences between bromodomains both within and across the different subfamilies, varies greatly.
The ATAD2B bromodomain is able to recognize acetylated lysine modifications on histone proteins (see below) [20]. The bromodomain is the only ATAD2B domain that has been studied, and only little information has been learned.
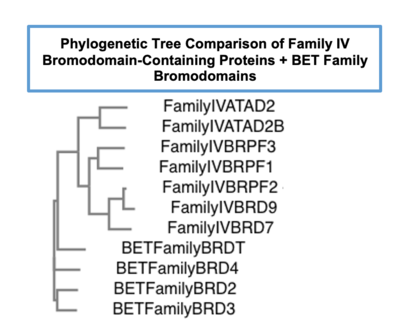 Phylogenetic tree comparing bromodomain-containing proteins in Family IV to the well characterized BET family of bromodomains | 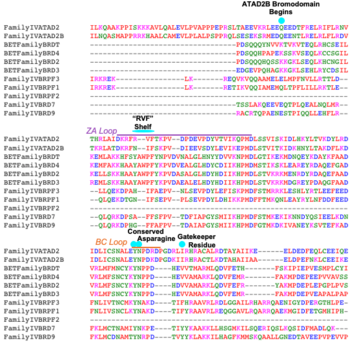 Sequence alignment for sub-families IV and BET (Family II), highlighting important structural characteristics of bromodomains |
Introduction into Bromodomain Structure
All bromodomain-containing proteins have the same structural motif. It is a left-handed four alpha-helical bundle. These alpha helices are connected by two different and flexible loops whose sequences are variable when compared to the other bromodomain residues, as seen above. These loops are responsible for coordinating the acetylated lysine histone residues to the bromodomain binding pocket. The binding pocket itself is hydrophobic, and for ATAD2 and ATAD2B, deep.
The is the single most important residue for bromodomain binding to acetylated lysine residues. Other residues that have been conserved throughout evolution, such as the preceding , are important for binding to the backbone of the histone tail. This works to help stabilize the acetylated lysine residue insertion into the bromodomain binding pocket. Additionally, there is a hydrophobic that serves to limit the number of acetylated lysine residues that are inserted into the binding pocket. Another motif in ATAD2 and other bromodomains, termed the "RVF" or "WPF" shelf also helps in ligand specificity in the ZA loop. ATAD2B has an "NIF" motif, which also helps it to distinguish between different histone ligands in Lloyd, et al. (2020)[21].
Interestingly, both ATAD2 and ATAD2B are known to recognize di-acetylated lysine residues on histone tails, but a structure of this interaction has yet to be solved. Unfortunately due to this, we are unable to visualize how both acetylated lysine residues fit in the binding pocket.
Bromodomain & ATAD2B Function
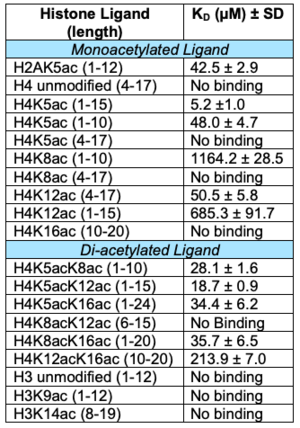
Binding affinities for the
ATAD2B bromodomain with histone ligands
Even though a structure doesn't exist with a ligand bound for ATAD2B, still coordinate interactions with the histone tails (Table). There is only one published manuscript detailing the histone binding partners for the ATAD2B bromodomain[22].
The bromodomain was able to distinguish between singly-acetylated lysine residues and di-acetylated lysine residues. When histone protein 4 (H4) was acetylated at lysine 5 (K5ac), the binding affinity was the tightest, at 5.2 µM and the weakest for H4K8ac at 1,164.2 µM. The ATAD2B bromodomain displays a 223-fold preference for the H4K5ac residue. ATAD2B also prefers the diacetylated ligand H4K5acK12ac, with a binding affinity of 18.7 µM, with 1.5-fold tighter binding than the next preferred residue H4K5acK8ac at 28.1 µM. ATAD2 also exhibits this same preference between these histone ligands. Interestingly, Koo, et al.[23] showed that the ATAD2 bromodomain recognizes that di-acetylated lysine modification (H4K5acK12ac) on nascent, or newly formed chromatin. Through recognition of the H4K5acK12ac modification on nascent chromatin/newly formed histones, ATAD2 is recruited to replication sites during chromatin organization and assembly. Even though the biological function of ATAD2 has never been fully elucidated, this manuscript brings a certain insight into its role in chromatin remodeling. While individual domains were not investigated in vitro, in vivo work displayed that when the AAA ATPase 1 domain and bromodomain was mutated, their ability to be replicated to nascent chromatin was lost. It is still unclear what specific role each domain has to play in ATAD2 function. ATAD2B has not been investigated in vivo, so it is yet to be determined if it is also recruited to newly synthesized histones. It is interesting that the acetylated lysine recognition trend is the same between the paralogs of ATAD2 and ATAD2B, but it is too soon to say if their functions are the same as well.
In Lloyd, et al.,[24] it was shown that mutation of the conserved asparagine residue (N1031A) severely weakened binding affinity to previously identified histone ligands. Mutagenesis of Y1037A, completely abolished binding affinity to previously identified histone ligand binding partners. Residues in the binding pocket of bromodomains, especially the ATAD2B bromodomain, are evolutionarily conserved and extremely important for interaction with the acetylated lysine histone ligands.
Detailed ATAD2/B Bromodomain Structure
Below are two images of the ATAD2B bromodomain to better investigate where the important binding residues are and their location to the histone ligand. The residues highlighted are able to coordinate binding to the epigenetic acetylated lysine PTMs on histone proteins. Unfortunately, there are no published structures of the ATAD2B bromodomain bound to any histone ligands. Since ATAD2 is a highly conserved paralog of ATAD2B, a structure of ATAD2 bound to a histone ligand containing the epigenetic PTM of acetylated lysine on histone tail residue 5 was aligned to the apo ATAD2B bromodomain structure, with an RMSD value of 0.608. The important binding site residues, especially those for the conserved asparagine, were indistinguishable. There was a slight variation between the preceding tyrosine residue, and more variation between the gatekeeper residues.
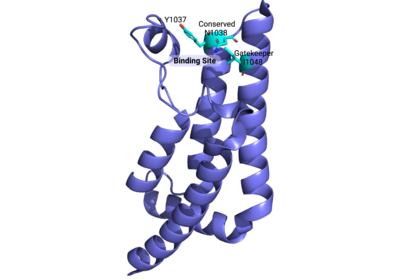 Important binding site residues highlighted above displayed here (cyan) in the ATAD2B bromodomain (PDBID: 3LXJ) | 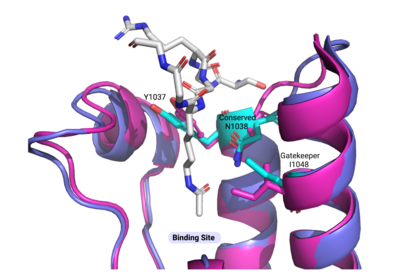 Alignment of the ATAD2B bromodomain (violet) with important binding site residues highlighted in cyan to the ATAD2 bromodomain (magenta) and the same important binding site residues. The RMSD value is 0.608, and there is little difference seen between these two paralogs and their important residues. |
But, ATAD2B also contains other domains, however no information about their function nor structure is available for them.
ATAD2/B Domain Organization:
Structural Visualization
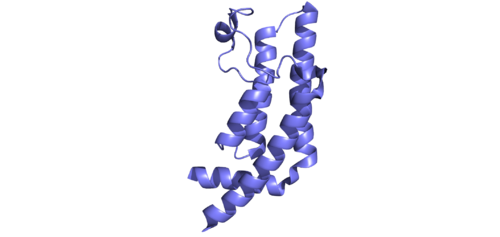
This domain organization is represented by the predicted structure of the ATAD2B protein using [25][26].
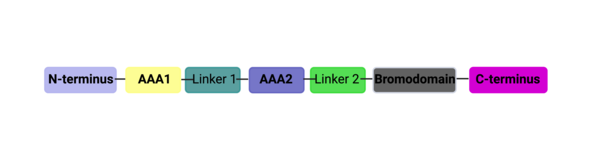
ATAD2B Domain Organization
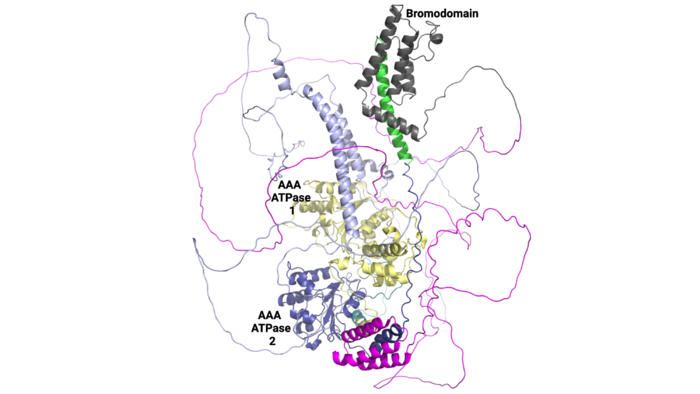
Domain Function
ATAD2 and ATAD2B are chromatin associated proteins (CAPs). Both the ATAD2 and ATAD2B proteins contain two conserved domains that can interact with chromatin: an and a [27]. Additionally, there are linker regions and a C-terminal domain, as represented by the AlphaFold-predicted structure. ATAD2 is a large protein (1,390 amino acids & 158.6 kDa) and has been studied primarily using cellular assays. In these assays, when both AAA ATPase and the bromodomain are present, ATAD2 has a tighter affinity for chromatin, which suggests both are important for the formation of regulatory chromatin complexes[28][29]. ATPase domains are known to promote the assembly of coregulator complexes at chromatin, and the ability of the bromodomain to recognize certain histone modifications may specify the location of where ATAD2 is recruited on chromatin[30]. Additionally, we know that ATAD2 is recruited to newly synthesized histones on nascent chromatin; moreover, ATAD2 is only present during the S-phase of the cell cycle [31], where chromatin remodelers are most active. Chromatin remodeler proteins are usually large complexes, and previously, it was determined that ATAD2 can oligomerize, and the first AAA ATPase is important for this oligomerization[32]. Recently, studies of ATAD2 using the yeast homolog Abo1, discovered that the ATPase domains formed a (using cryo-EM) and was involved with the loading of H3 and H4 onto the NCP. The C-terminal domain was necessary for hexamer stability. However, the bromodomain could not be visualized using cryo-EM. Another yeast homolog, Yta7, found ATAD2 to function as a histone chaperone. The in-depth molecular functions of ATAD2, and ATAD2B still remain incredible elusive despite recent advances, due to this lack of structural information.
ATAD2B has not been extensively studied in the same way as ATAD2, so even less information on structure and function are available.
While the AlphaFold structure gives a nice idea of what the full structure of ATAD2B looks like, not enough is known about the overall function of ATAD2B. More information is greatly needed in order to learn more about its role in chromatin remodeling, oncogenesis, development, and overall place in the cell.
Therapeutic Interventions
Many bromodomain-containing proteins are highly overexpressed in many forms of cancer, and the pharmaceutical industry is keen to design inhibitors to temper bromodomain function in cancer progression. The most well-known inhibitor is a pan-bromodomain inhibitor, JQ1 that was originally designed for the BET Family bromodomains. Over time, it has been characterized to have slightly inhibitory effects in other bromodomains. ATAD2B does not have a specific inhibitor, but ATAD2 has a few currently commercially available. The ATAD2 bromodomain, however, is notoriously extremely difficult to drug, due to the deepness of its binding pocket[33]. Only a handful of the compounds developed have been shown to be effective in blocking the binding interaction between ATAD2 and acetylated lysine PTMs[34][35][36]. Despite ATAD2B not having its own selective inhibitor, Llloyd, et al.[37] characterized the effects of a known ATAD2 inhibitor, Compound 38, with the ATAD2B bromodomain.
Compound 38
Compound 38 (C-38) was 1.8-fold times more selective for the ATAD2 bromodomain than ATAD2B, with 90 nM and 166.3 nM binding affinities, respectively. They solved the of C-38 bound to the ATAD2B bromodomain to determine the specific molecular interactions that are occurring (PDB ID: 6VEO). C-38 was coordinated in the binding pocket much like the acetylated lysine residues of the histone proteins: through hydrogen bonds and hydrophobic interactions. The inhibitor bound to the conserved asparagine (N1038), and was coordinated through hydrophobic interactions with the gatekeeper residue I1048. However, since this compound was manufactured to be specific for the ATAD2 bromodomain, some important interactions between protein and inhibitor are not present in the ATAD2B bromodomain structure, including an important hydrogen bond interaction. These mechanistic molecular differences are thought to play a role in the lower affinity for C-38 with ATAD2B. Overall, however, the coordination of C-38 between ATAD2 and ATAD2B is extremely similar, and Lloyd, et al.[38] postulates that it may be difficult to develop a truly selective inhibitor for each protein. This endeavor is still being explored.
Conclusion
Overall, more information is needed in order to understand the true biological function of ATAD2B. Due to bromodomains being conserved throughout evolution and after being divided into eight sub-families, it is fortunate to have similar proteins to study the structure and function of ATAD2B through. We know that the bromodomain can recognize mono- and di-acetylated histone proteins with micromolar affinity, and that the most important binding residues align well with the ATAD2 bromodomain, despite there not being a structure for ATAD2B bound to a histone protein.
AlphaFold helps us to visualize the entire length of the ATAD2B protein. While interesting and insightful, there are many loop regions, and we do not know what the function of the other domains are yet. It would be interesting to investigate ATAD2B further to determine more about its function so that we can learn from its structure.
There are many players in the story and function of ATAD2B, but little knowledge. In the future, it will be interesting to see what new insights develop into its structure and function.
Additional Resources
ATAD2 UniProt Page
ATAD2B UniProt Page
PubMed Information
EpiCypher Ways to decipher the epigenetic landscape and its interacting partners
Acknowledgments
Glass & Frietze Laboratories : UVM researchers working on learning more about the function of ATAD2, and especially ATAD2B!
Images created with BioRender.com & PyMOL Molecular Graphics System