User:Melanie Ng/Sandbox 1
From Proteopedia
Mutant F99S/M153T/V163A
|
Three mutations on the beta barrel of the Green Fluorescent Protein, F99S, M153T and V163A, causes this specific mutant to fluoresce much greater than the wild type protein. As a result it serves as a better marker for gene expression in vivo.
Contents |
Background
Green Fluorescent Protein (GFP) was acquired from the luminescent Aequorea victoria jellyfish. Wild type GFP is a 238 amino acid protein and the fluorescent choromophore is formed by an internal cyclization of a Serine-Tyrosine-Glycine tripeptide. GFP absorbs blue light primarily at 395 nm accompanied by a smaller peak at 475 nm and emits green light at 509 nm with a smaller peak at 540 nm. The Green Fluorescent Protein has been used as a reporter protein for gene expression, protein localization and cell linage information. The slow rate of fluorescence attainment in vivo limits the GFP as a reporter for gene expression. Considerable variations of GFP have been created in order to help determine the structure and function of the wtGFP. These mutations have been made in order to enhance the use of GFP as a reporter of gene regulation and expression, protein transport, and heredity. GFP has been mutated to provide an increased or alternative color for fluorescence, for a greater time period and to make it less susceptible to photo bleaching. The specific GFP mutation model featured is referred to as Mutant F99S/M153T/V163A or cycle 3 GFP (c3-GFP).
Structural Properties
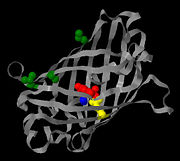
This protein, as its name implies, is mutated at the 99 residue site where the amino acid Phenylalanine is replaced with Serine (), the 153 residue site where Methionine is replaced by Threonine ( ), and the 163 residue site where Valine is replaced with Alanine (). Each mutation site of the c3-GFP is located near the surface of the protein and on separate beta sheets. In contrast to many mutations that enhance fluorescence, such as the S65T, which have the mutant residue proteins in close proximity to the internal chromophore and directly participating in conformational changes of the chromophore; the mutant residues of c3-GFP are not in close proximity to the chromophore and subsequently do not directly interfere with the normal cyclization and formation of the (red molecule). , colored yellow and two water molecules colored blue are also featured in this model. The glutamate is present because it is one of the most conserved residues in GFP. Two water molecules are included in this model to represent the A and B chains of the c3-GFP, while the C and D chains, which are not represented, contain three water molecules.
Chemical Properties
The overall characteristic of the c3-GFP closely resemble the wtGFP, however due to the three mutations it is 42-fold more fluorescent in vivo compared to wtGFP. This phenomenon arises because the replacement of the hydrophobic amino acids with hydrophilic or less hydrophobic amino acids causes less aggregation and results in improved autocatalytic activation of the chromophore. The reduction in the hydrophobic nature on the surface of the c3-GFP also results in an increased ability of the mutant to mature in vivo effectively at 370C. This is a feature the wtGFP lacks because the A. victoria jellyfish in the Pacific Northwest from which it was first extracted had never experienced such high temperatures, thus the wtGFP has its limitations as a gene expression marker. The c3-GFP mutant is special because unlike most mutants, the structure of the chromophore and the fluorescence spectrum remains the same as the wtGFP. The enhanced GFP provides for a better tool for researchers as compared with the wtGFP.
References
1. Battistutta R, Negro A, Zanotti G. 2000. Crystal Structure and Refolding Properties of the Mutant F99S/M153T/V163A of the Green Fluorescent Protein. Proteins: Structure, Function, and Genetics 41:429 – 437.
2. Crameri A, Whitehorn E, Tate E, Stemmer W. 1996. Improved Green Fluorescent Protein by Molecular Evolution Using DNA Shuffling. Nature Biotechnology 14:315 - 319.
3. Fukuda H, Arai M, Kuwajima K. 2000. Folding of Green Fluorescent Protein and the Cycle3 Mutant. Biochemistry 39:12025 – 12032.
4. Martin Chalfie, et al. 1994. Green Fluorescent Protein as a Marker for Gene Expression. Science: New Series 263:802-805.