User:Michael Adams/Sandbox 1
From Proteopedia
|
Contents |
Arginine Kinase
A phosphokinase used to store energy in the form of Argininephosphate.
Isolation
In Strong and Ellington’s 1994 experiment, arginine kinase (AK) was isolated from Limulus polyphemus, the Atlantic horseshoe crab, a marine chelicerate arthropod. They isolated the gene for AK, sequenced the DNA and identified it to be 1071 nucleotides. The 1071 nucleotides produce a 357 amino acid protein and AK extracted from other organisms show similarity to this protein. AK serves a similar function to that of creatine kinase, in vertebrates [1]. The enzyme creatine kinase maintains energy homeostasis by producing ATP in high energy requiring cells such as skeletal and cardiac muscle and neurons [2]. This is done by the transfer of an N-phosphoryl group from phosphocreatine to ADP.
Structure
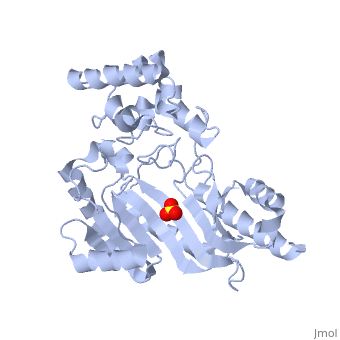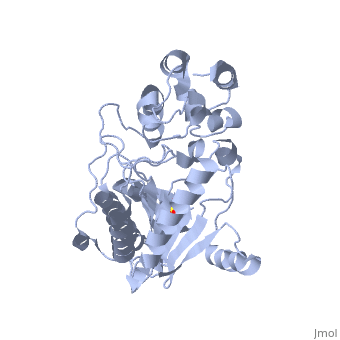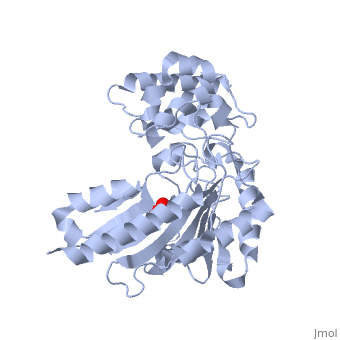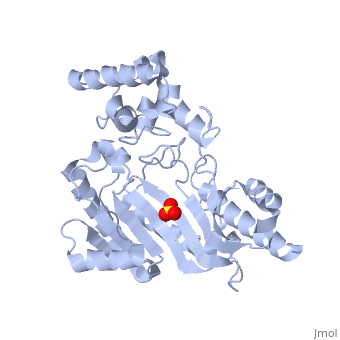
The structure of is mainly α-helical and contains an N-terminal region with a specificity loop for specific substrate binding. (Figure 2). However, when compared to creatine kinase, arginine kinase is not terminated at the N-terminal end with a pair of proline-glycine residues. Typically within creatine kinase, the proline molecules restrict changes in conformation and is the amino acid that terminates helices. The glycine chains are usually associated with flexibility. However, in arginine kinase this is typically not the case. On the C-terminal end, there is an eight-stranded antiparallel with seven flanking the sheet (Figure 1). Residue is an arginine that appears to play a crucial role in maintaining structural stability. Studies show that a mutation in the residue leads to a steep decline in enzymatic activity [3].
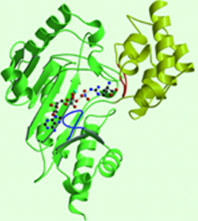
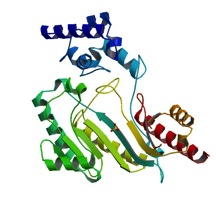
The small domain specificity loop forms a “specificity” pocket surrounding the methyl substituent of the guanidinium group that is unique to creatine substrates. In this region, five residues differ between arginine and creatine kinases: 312, 314, 315, 317, and 319 [6]. Within each arginine kinase, there is typically a Mg+2 ion adjacent to the antiparallel β-sheet (Figure 2) to aid in the increase of ATP’s affinity for the binding site [7]. Typically two arginine kinase structures mirror each other and form a hole like structure in between the two (Figure 1).
Function
Arginine Kinase is part of a class of kinases that regulates ATP levels in the body to help maintain homeostasis. Arginine Kinase is a phosphokinase - a kinase used in the catalyzation of phosphagens and adenosine diphosphate (ADP) into adenosine triphosphate (ATP). It creates a sort of storage option for energy. Phosphagens act as a storage form of phosphate (Nω-phospho-L-arginine) that can be catalyzed into an energy source (ATP) when needed [8]. The arginine kinase is a lock and key catalyst that holds ADP and phosphoarginine in place and catalyzes the transfer of phosphate from phosphoarginine to ADP forming arginine and ATP [9]. It is the most common phosphokinase (PK) in invertebrates. The most common PK in vertebrates is Creatine Kinase [8]
Application to the Animal Kingdom
Arginine Kinase is the individual phosphagen kinase that is found in major invertebrates, such as: arthropods, mollusks, and echinoderms. Most recently, an arginine kinase was purified from a house fly [2]. This gave Wallimann and Eppenberger the initiative to investigate the arginine kinase in Drosophila melanogaster, also known as a fruit fly [2]. Since the genome and genetic development of Drosophila melanogaster is well understood, this allows for any discoveries made to be easily interpreted. Additionally, further discoveries will help better understand the characteristics of arginine kinase corresponding vertebrate enzyme, creatine kinase [2].
Arginine Kinase (AK) is represented by a single gene and the sequence or partial sequence is available from Drosophila, Limulus, lobster, shrimp, and abalone [10]. In the study by Wang et al. 1998, a phylogenetic tree was put together for the evolution of Arginine Kinase compared to Creatine Kinase. It indicates that there are major clusters corresponding to Creatine Kinase and Arginine Kinase. Also, it is expressed in the gills of two species of euryhaline crabs, the blue crab Callinectes sapidus and the shore crab Carcinus maenus, in which energy-requiring functions include monovalent ion transport, acid-base balance, nitrogen excretion [11].
References
- ↑ Strong, S., & Ellington, W. (1994). Isolation and sequence analysis of the gene for arginine kinase from the chelicerate arthropod, Limulus polyphemus: Insights into catalytically important residues. Biochimica Et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology, 197-200.
- ↑ 2.0 2.1 2.2 2.3 Wallimann, Theo, and Hans M. Eppenberger. "Properties of Arginine Kinase from Drosophila Melanogaster." Www.onlinelibrary.wiley.com. Laboratory of Developmental Biology, Swiss Federal Institute of Technology, Zurich, 21 June 1973. Web. 12 Nov. 2015
- ↑ Wang WD, Wang JS, Shi YL, Zhang XC, Pan JC, Zou GL. Mutation of residue arginine 330 of arginine kinase results in the generation of the oxidized form more susceptible. Int J Biol Macromol. 2013 Mar;54:238-43. doi: 10.1016/j.ijbiomac.2012.12.015., Epub 2012 Dec 19. PMID:23262386 doi:http://dx.doi.org/10.1016/j.ijbiomac.2012.12.015
- ↑

- ↑ http://www.rcsb.org/pdb/explore.do?structureId=4GVZ
- ↑ Newsholme, E. A., Beis, I., Leech, A. R., & Zammit, V. A. (1978). The role of creatine kinase and arginine kinase in muscle. Biochemical Journal, 172(3), 533–537.
- ↑ doi: https://dx.doi.org/10.1080/08927022.2011.561430
- ↑ 8.0 8.1 Pereira, C. A., Alonso, G. D., Paveto, M. C., Iribarren, A., Cabanas, M. L., Torres, H. N. & Flawia, M. M. (2000) Trypanosoma cruzi arginine kinase characterization and cloning., J. Biol.Chem. 275, 1495-1501.
- ↑ Azzi A, Clark SA, Ellington WR, Chapman MS. The role of phosphagen specificity loops in arginine kinase. Protein Sci. 2004 Mar;13(3):575-85. PMID:14978299 doi:10.1110/ps.03428304
- ↑ Wang, Yu-mei E., Pia Esbensen, and David Bentley. "Arginine Kinase Expression and Localization in Growth Cone Migration." The Journal of Neuroscience 18.3 (1998): 987-98. Web. 15 Nov. 2015. <http://www.jneurosci.org/content/18/3/987.full.pdf>
- ↑ Kotlyar, S., Weihrauch, D., Paulsen, R., & Towle, D. (2000, July 20). Expression of Arginine Kinase Enzymatic Activity and mRNA in Gills of the Euryhaline Crabs Carcinus Maenas and Callinectes Sapidus. Retrieved November 17, 2015, from http://jeb.biologists.org/content/jexbio/203/16/2395.full.pdf