User:Nikhita Khanna/Glucosamine 6-Phosphate
From Proteopedia
|
Contents |
ABSTRACT
C. albicans is a fungus normally present on the skin and within the mucous membrane. Despite its common presence, any stimuli for overgrowth can induce the invasion of C. albicans into the throat, intestines, and heart valves as it travels down the bloodstream. The fungus can be present in various morphologies such as rounded buds (yeast), pseudohyphae, and hyphae (mycelia). However, it is the hyphal form that induces the invasion into tissue. Glucosamine 6 phosphate (GlcNAc) synthase is an enzyme that catalyzes the first step in the pathway that ultimately results in hyphal formation. As a result, UDP-GlcNAc, a precursor of chitin, is generated. However, the specific mechanisms for the production of UDP-GlcNAc by this enzyme are not clearly understood.
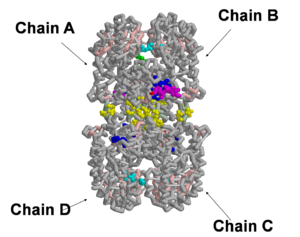
Eukaryotic GlcNAc synthase is a dimer of two individual dimers, which then combines to display a tetrameric structure. Each subunit is composed of two domains, GAH and ISOM. GAH is involved in glutamine hydrolysis while the ISOM domain is directly involved with the isomerization of fructose 6 phosphate to glucose 6 phosphate. We have analyzed and constructed a 3D physical model of GlcNAc synthase that focuses on the ISOM domain (amino acids 346-712) in complex with UDP-GlcNAc and fructose 6 phosphate using the computer software RasMol. The secondary structure of each subunit involves beta sheets (light pink) with hydrogen bonds (white) that provide stabilization to the model. In addition, we have selected residues involved in the tetramerization of this enzyme, an intermolecular interaction that is not observed in prokaryotes. Our physical model depicts residues involved in tetramerization (aa 524-527). Other contacts related to tetramerization between the subunits are present in residues 391-445 (yellow). We have also selected fructose 6 phosphate (light green) in complex with the enzyme. Residues of the enzyme that are in close proximity to this molecule; Glu591, Lys588, His607 (light blue) reveal a possible binding region. Since the experimental procedures involved the replacement of GlcN-6P with Fructose-6P, and the combination of the obtained crystal structures with and without Fructose-6P using computer software, we only see the presence of one Fructose-6P ligand binding despite four possible binding sites. Finally, UDP-GlcNAc (magenta) molecule and surrounding amino acids 474-492 (royal blue) are represented to highlight the binding pocket. Although UDP-GlcNAc is present in complex only on chains A and B, the binding pocket is evident on all four chains. This is due to the fact that there were two crystal structures obtained for the synthase. Crystals were grown both in the absence of UDP-GlcNAc and in the presence of UDP-GlcNAc (Konariev et all 2007). Both crystal structures were used to manually form the final model within our referred PDB structure (2PUT) using computer software, and thus we see the UDP-GlcNAc binding in one chain, while the other dimer chain of each pair is free of the ligand.
Although the distinct tetramerization, fructose interaction sites, and UDP-GlcNAc binding pocket selected in the physical model represent important areas in the development of the invasive mycelia form of C. albicans, there are numerous other factors such as phosphorylation sites present on the GAH domain, amino acids involved in amido transfer, and other regions which are also highly significant. Further investigation could reveal possible competitive inhibition for the binding region of fructose and tetramerization sites which can the synthase and thus prevent the formation of UDP-GlcNAc and ultimately inhibiting mycelia transition. As we further explore the structure of the synthase and other molecules in the pathway, we can further elucidate such mechanisms.
C. Albicans as a Pathogen – Yeast to Mycelia Candidiasisis a mycosis (fungal infection) that encompasses minor rashes to life threatening situations, especially in Immunocompromisedpatients. It is most commonly caused by the opportunistic pathogen Candida Albicans, a fungus commonly found on our skin and mucous membrane. The transition of the fungus to the pathogenic form is induced by environmental conditions and secondary messengers that initiate a corresponding transition from the yeast to mycelial (hyphae) form. It is the mycelial form is associated directly with invasion into tissue. In order to make the metamorphosis into mycelia, it is essential to activate the molecular pathways that lead to the formation of chitin, as a higher chitin content is necessary in the cell wall of hyphae. The first pathway that is initiated results in the synthesis of UDP-GlcNAc, a precursor of Chitin.
GlcN6P-Synthase in the mycelial transformation The metabolic pathway involves four steps, each catalyzed by a different enzyme. Glucose-amine-6-Phosphate synthase catalyzes the first step of the pathway resulting in the conversion of Fructose-6-Phosphate to Glucosamine-6-Phosphate The conversion itself involves two steps; glutamine hydrolysis and the isomerisation of Fructose-6-Phosphate. Both steps are correlated directly with the two separate domains of the synthase respectivelythe N-terminus GAH and C -terminus ISOM. The exact mechanism of the synthase activity is not fully understood, but there are quite a few areas of significance to the actual mechanism of catalysis by the synthase. The interaction between the two domains is related to a hydrophobic channel that ensures sufficient ammonia transfer between the two, and there are various sites, especially those related to phosphorylation (an inducer of greater chitin content) within the GAH domain. Although the action of the enzyme involves the dynamic use of the entire structure, it is the ISOM domain that participates directly in the binding of ligands such as GlcN-6P, Fructose-6P, and the negative feedback inhibitor UDP-GlcNAc. We have examined the structure and functional mechanisms of the ISOM domain in detail and deduced some of the vital sites of ligand interaction and interaction within the protein structure itself that might pose value in the full knowledge and understanding of its metabolic pathway.
METHODS AND RESULTS
The Protein Data Bank (PDB) was used as a source of the model with the following PDB ID: 2PUT [The Crystal Structure of Isomerase Domain of Glucosamine-6-Phosphate Synthase from Candida Albicans].
RasMol was used to highlight the important features in the model. The following values were used to display the model for optimal viewing:
- Backbone 300
- Spacefill 275
- Monitor lines 200
- Hydrogen bonds 225
DISCUSSION
|
A 3-dimensional model of Glucosamine-6-phosphate synthase was created highlighting vital features of the protein’s isomerase domain:
- The includes the sidechains Gly474, Val476, Ser484, Thr487, His492 and residues 489-491.
- Coordinates with when is bound, possibly indicating that a positive charge is needed to increase the stability of the enzyme-substrate complex.
- interacts with of Glu591, Lys588, His607. These residues form a small binding domain for F6P, which has yet to be completely characterized.
- These two important binding sites appear on opposite sides of each chain, perhaps indicating that the substrates do not greatly effect each others’ interaction with the protein.
- create a horizontal line of symmetry across the protein, separating identical subunits (A and D, B and C), a so-called “dimer of dimers.”
- could also provide internal support to the protein.
A physical model of the isomerase domain, the main ligand binding structure of the protein, can aid in the understanding of its multiple substrate interactions and necessary conformations for enzymatic activity. With this new tool, we can propose ways to inhibit substrate binding, and consequently, the metabolic pathway of C. Albicans.
FUTURE GOALS
Recently, it has been determined that the cAMP signaling pathway in Candida Albicans is the central signaling pathway essential for hyphal morphogenesis. However, the molecular mechanism responsible for regulating this pathway has not been clearly understood.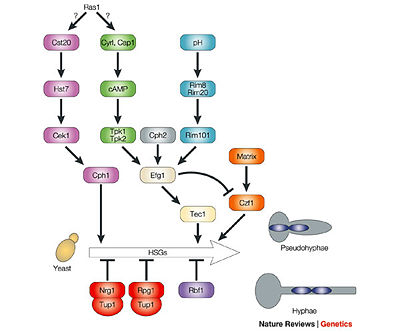
Shown above is the Ras1-cAMP-Efg1 signaling cascade which is an important factor in the hyphal morphogenesis of the Candida Albicans. (Berman)
Cdc35 is a single gene in C. albicans that is homologous to Saccharomyces cerevisiae adenylyl cyclase gene. This cyclase is not essential for growth in Candida albicans, however it is required for hyphal development. It has been observed that the CaCdc35 protein at the plasma membrane is indicated by a weak signal, meaning that CaCdc35 is recruited to the membrane, but its exact mechanism of association with the membrane is unknown.
There are several other paths that can be taken in order to better understand Candida albicans and its hyphal formation. The model of Candida albicans printed by our team can be used to visualize the physical structure and position of the Glucosamine 6-phosphate (GlcN-6-P) synthase. The GlcN-6-P synthase ia an enzyme that catalyzes the first step in the reaction pathway leading to the formation of uridine 5'-diphosphate-N-acetyl-D-glucosamine (UDP-GlcNAc), a precursor of macromolecules that contains amino sugars. If, with the help of more advanced computer software, changes in the physical structure of the GlcN-6-P synthase can be viewed upon the binding of the fructose 6 phosphate, then it can help determine what factors are responsible for the hyphal formation of Candida albicans.
Since only a few residues of the binding site of the fructose 6 phosphate are known, the potential interactions between fructose 6 phosphate and the surrounding binding region may lead to the determination of the entire binding pocket. Another major aspect of the structure that was printed was the presence of the sodium ions. It has not clearly been determined why they are important to this structure, but somehow they play a significant role in the stabilization of the binding sites located throughout the GlcN-6P synthase.
Throughout many studies, it has been noticed that one technical problem in the analysis of Candida albicans is the detection of the GFP (Green Fluorescent Protein) tagged proteins. The signal for GFP detection through microscopy is shown to be weak in past experiment, which prevents the study of GFP regulation. In that sense, a better folding GFP protein can help. The signal strength of GFP can be improved by using different GFP variants in Candida Albicans.
The two important signaling pathways that activate CaCdc35 in Candida albicans to stimulate the hyphal development are Cdc25-Ras 1 and Gpr1-Gpa2. Gpa2, which encodes an alpha subunit homologue of the heterotrimeris G-protein, was proposed to function in the regulation of cAMP levels. Gpr1 encodes a protein that is predicted to contain seven membrane-spanning helices, a feature characteristic of G protein-coupled receptors. It has been confirmed that, the Gpr1 protein is an important plasma membrane sensor, and its deletion produces defects in hyphal formation and morphogenesis in Candida albicans. It has also been known that the adenylyl cyclase CaCdc35, which is involved in CO2 sensing, is directly interacts with Ras1, the single Ras homologue in C. albicans, but its interaction mechanism with Gpa2 is unknown. Mutants lacking Ras1 are viable but have a severe defect in hyphal growth in response to serum and other inducers.
Another major question arises with the identity of the factors that activate CaCdc25- the guanine nucleotide exchange factor (GEF) that activates Ras1. Ligands of the plasma membrane sensor CaGpr1 are also not characterized well, although amino acids, especially methionine, are thought to be one potential candidate.
Structural studies of the plasma membrane receptors and the critical components of the signal pathway will have to be addressed in the near future. And this will help in the design of new inhibitors of Candida albicans signal pathways and also for developing new strategies of antifungal therapies.
References
- Berman, Judith, and Peter E. Sudbery. 2002. Candida albicans: A molecular revolution built on lessons from budding yeast. Nature Reviews Genetics 3:918-932. <
 >
>
- http://dspace.sunyconnect.suny.edu/bitstream/1951/44897/1/105531432.sbu.pdf