User:Olivia Cheng/Sandbox 1
From Proteopedia
Outer Surface Protein B (OspB) has been found to play a vital role in the adherence of B. bugdorferi onto tick guts, which promote the survival of the vector and spread of Lyme disease. OspB-deficient B. budgdorferi have been found to bind poorly to tick gut extracts. The expression of OspB, along with OspA, is upregulated and downregulated by B. Burgdorferi according to the distinct phase of the life cycle that it is in. When the spirochete resides inside the arthropod vector, OspB is upregulated to promote binding to the tick’s gut. However, during transmission from the tick to a vertebrate host, OspB is downregulated and other proteins such as OspC, DpbA and BBK32 are upregulated [1].
Contents |
Background
|
Lyme disease, discovered in 1975, is spread by means of a tick vector that carries the causative agent, spirochete Borrelia Burgdorferi [2]. It is the most frequently reported vectorborne illness in the United States. Individuals who have this disease may show symptoms including skin lesions called erythema migans and the characteristic bulls-eye rash [3]. OspA and OspB are the major outer surface proteins found on B. Borrelia. OspB has shown significant variability in amino acid sequence and antigen reactivity in comparison to OspA, known to be largely invariant [4].
OspB Interaction with Fab of H6831
The outer-surface proteins (Osps) in B. burgdorferi spirochete activate the classical and alternative pathways of the complement system. B. burgdorferi is resistant to complement mediated lysis. The complement inhibitor factor H binds to Osps and the C3b cascade is deactivated [4].
Within this complex is , shown in purple. The H6831 Fab is shown in white. (Click to revert back to the original.)
H6831 is an IgG class monoclonal complement-independent antibody shown to effectively lyse outer surface protein B (OspB) of B. burgdorferi. H6831 recognizes on OspB. Studies have shown that B. burgdorferi strains with Thr, Cys, Gly, or Glu instead of Lys decrease the binding affinity between H6831 and OspB. The sequence and structure of bactericidal H6831 Fab are typical for IgG2 heavy chain/kappa light chain class. [4].
The H6831 epitope of OspB is topologically equivalent to LA-2 epitope of OspA [4]. Similar to the LA-2 epitope, the H6831 epitope is positioned opposite the N-terminus near the end of the antigen. The buried surface area of OspB in the H6831 Fab complex is smaller than that of the OspA-LA2 complex. Loop 1 in the OspA-LA2 complex has the most interactions with the Fab, where as Loop 1 in the OspB-H6831 complex has the fewest interactions with the Fab [5].
Binding
|
H6831 Fab complex binds to a highly accessible region near the C-terminus of OspB, away from the N-terminus lipid anchor. The interaction of OspB-Fab complex depends heavily on hydrogen bonding between . When binding of full-length OspB to Fab fragments of H6831 or CB2 fail, it is usually because , located on loop 2, has been replaced with a different residue. Studies show that substitutions in basic residues of hen egg-white lysozyme (HEL) that participate in HEL-Fab complexes decreased binding affinity by 400-10,000 times. [4]
Due to its effective bactericidal actions, H6831 is used to generate less virulent escape variants of B. burgdorferi [4]. In the majority of the mutations created from in vivo and in vitro immunization of mice, truncated forms of OspB within the C terminus lead to premature stop codons[6]. It has been suggested that OspB mutants are more sensitive to proteolysis due to missense mutations that disturb the conformation of OspB [4]. Truncated OspBs cease within the two C-terminal beta-strands of the central sheet. H6831 disorders or removes a beta sheet from OspB after binding. Cleavage may be a possible explanation for the conformational changes of OspB [7]. In and forms of OspB, some changes result from proteolysis near the N terminus [4]. Residues 157 - 201 on OspB contain the , shown in plum.
Aromatic residues tyrosine and tryptophan are also present in the OspB-H6831 interaction, a feature found in many antigen-antibody complexes. The Lys-253 residue forms a trans conformation between these aromatic residues of H6831. In the complex structure of the antibody binding site, the electron density is well defined and shows increased contact between Lys-253 and the antigen-binding site of the Fab. Most of the electrostatic and hydrogen-bond interactions occur between loop 2 and the Fab heavy chain [2].
Potential Mechanisms of Lysis
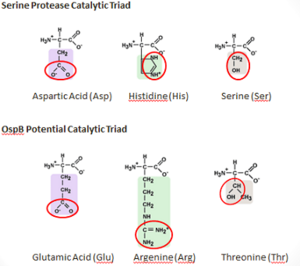
Potential Catalytic Triad
|
The mechanism by which H6831 Fab destroys a spirochete appears to be a novel interaction. It is possible that Fab binding changes the properties of OspB folding, which may increase sensitivity of the protein to proteolysis or aggregation. NMR methods showed that the effects of binding can be sent to regions of the antigen distant from epitope, which is at the shown in red (N-terminus in blue). OspB shows signs of truncation after interacting with Fab of H6831 [8].
It is possible that OspB performs an autoproteolysis. There is a found on OspB that resembles the catalytic triad of Serine_Proteases. This "constellation" consists of Thr-166, Arg-162, and Glu-184, which is similar to the catalytic triad residues of the serine protease trypsin, which are Ser-195, His-57, Asp-102 [9].
Threonine and Glutamic acid are found in other catalytic triads of the serine hydrolase family, but argenine seems unlikely to replace histidine as a base because of its higher pKa. There have been studies that have shown that Argenine is essential for other enzymatic functions, such as in the Ser-Arg-Asp triad in cytosolic phospholipase A2 and as a catalytic base in Sortase A. forms an H-bond with and may rearrange to form a putative oxyanion hole with Thr-166 and another unidentified atom if active in the catalysis. A concerted proton transfer, similar to a “proton wire”, is one plausible mechanism that would allow argenine to function in the catalytic triad of a protease [2].
Potential Oxidative Mechanism
It was recently discovered that all antibodies contained Fab portions that catalyzed a reaction between singlet oxygen and water, yielding hydrogen peroxide, ozone, water and hydroxide radicals. Hydrogen peroxide is a toxic oxidative species and might be the product of an ancient mechanism to protect against infection. UV absorption increases the rate for this reaction. B. burgdorferi is especially vulnerable to oxidative damage because its ecological niche is in areas with limited oxygen and its genome does not encode a catalase. This oxidative mechanism might explain why some mABs are bactericidal without the use of complement [2].
3D Structures
Links
- cdc.gov/lyme/ - the Official Lyme Disease resource of the US Center for Disease Control.
References
- ↑ Neelakanta G, Li X, Pal U, Liu X, Beck DS, DePonte K, Fish D, Kantor FS, Fikrig E. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog. 2007 Mar;3(3):e33. PMID:17352535 doi:10.1371/journal.ppat.0030033
- ↑ 2.0 2.1 2.2 2.3 Connolly SE, Benach JL. The versatile roles of antibodies in Borrelia infections. Nat Rev Microbiol. 2005 May;3(5):411-20. PMID:15864264 doi:10.1038/nrmicro1149
- ↑ http://www.cdc.gov/lyme/stats/index.html
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 Becker M, Bunikis J, Lade BD, Dunn JJ, Barbour AG, Lawson CL. Structural investigation of Borrelia burgdorferi OspB, a bactericidal Fab target. J Biol Chem. 2005 Apr 29;280(17):17363-70. Epub 2005 Feb 15. PMID:15713683 doi:10.1074/jbc.M412842200
- ↑ Golde WT, Piesman J, Dolan MC, Kramer M, Hauser P, Lobet Y, Capiau C, Desmons P, Voet P, Dearwester D, Frantz JC. Reactivity with a specific epitope of outer surface protein A predicts protection from infection with the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1997 Mar;65(3):882-9. PMID:9038292
- ↑ Schwan TG, Schrumpf ME, Karstens RH, Clover JR, Wong J, Daugherty M, Struthers M, Rosa PA. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J Clin Microbiol. 1993 Dec;31(12):3096-108. PMID:8308101
- ↑ Escudero R, Halluska ML, Backenson PB, Coleman JL, Benach JL. Characterization of the physiological requirements for the bactericidal effects of a monoclonal antibody to OspB of Borrelia burgdorferi by confocal microscopy. Infect Immun. 1997 May;65(5):1908-15. PMID:9125579
- ↑ Benjamin DC, Williams DC Jr, Smith-Gill SJ, Rule GS. Long-range changes in a protein antigen due to antigen-antibody interaction. Biochemistry. 1992 Oct 13;31(40):9539-45. PMID:1382591
- ↑ Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002 Dec;102(12):4501-24. PMID:12475199