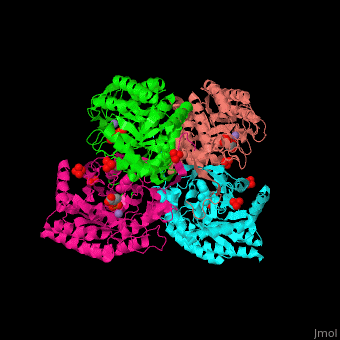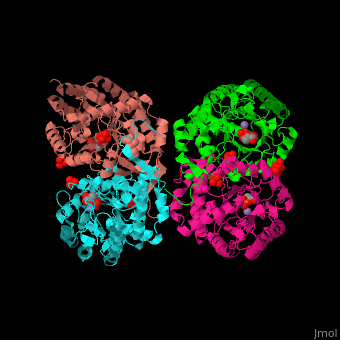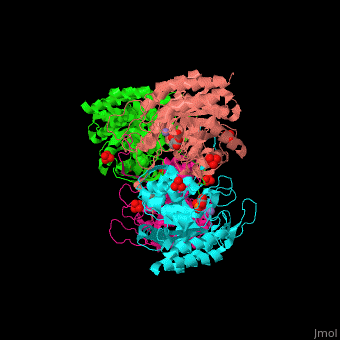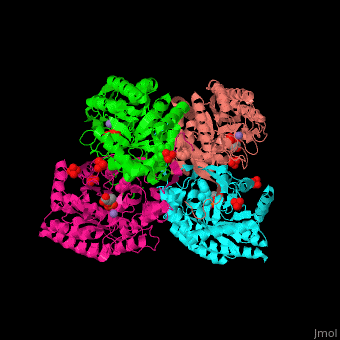
DAHP synthase
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='E. coli DAHP synthase complex with PEP, phenylalanine, sulfate and Mn+2 ion (PDB code [[1kfl]])' scene='70/708806/Cv/1'> |
== Function == | == Function == | ||
| - | '''DAHP synthase''' or '''3-deoxy-D-arabino-heptulosonate 7-phosphate synthase''' (DAHPS) catalyzes the conversion of phosphoenolpyruvate (PEP) and D-erythrose 4-phosphate to 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) and phosphate. DAHPS is part of the shikimate pathway. DAHPS requires a bivalent metal ion cofactor for normal activity. | + | '''DAHP synthase''' or '''3-deoxy-D-arabino-heptulosonate 7-phosphate synthase''' or '''phospho-2-dehydro-3-deoxyheptonate aldolase''' (DAHPS) catalyzes the conversion of phosphoenolpyruvate (PEP) and D-erythrose 4-phosphate to 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) and phosphate. DAHPS is part of the shikimate pathway. DAHPS requires a bivalent metal ion cofactor for normal activity. <scene name='70/708806/Cv/6'>DAHPS is a tetramer</scene>. DAHPS exhibits feedback inhibition by aromatic amino acids like tyrosine, phenylalanine and tryptophan.<ref>PMID:1682314</ref> |
== Structural highlights == | == Structural highlights == | ||
| - | + | The DAHPS active site is located in a channel at the C-terminal of the enzyme where the <scene name='70/708806/Cv/7'>substrate (PEP)</scene>, <scene name='70/708806/Cv/8'>inhibitor (phenylalanine)</scene> and <scene name='70/708806/Cv/9'>metal ion (Mn+2)</scene> are seen. The bivalent metal is bound to a Cys-X-X-His motif.<ref>PMID:12126632</ref> | |
| - | + | ||
| - | + | ||
== 3D Structures of DAHP synthase == | == 3D Structures of DAHP synthase == | ||
| + | [[DAHP synthase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Stephens CM, Bauerle R. Analysis of the metal requirement of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Escherichia coli. J Biol Chem. 1991 Nov 5;266(31):20810-7. PMID:1682314
- ↑ Shumilin IA, Zhao C, Bauerle R, Kretsinger RH. Allosteric inhibition of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase alters the coordination of both substrates. J Mol Biol. 2002 Jul 26;320(5):1147-56. PMID:12126632