We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Q165E/S254K Double Mutant Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
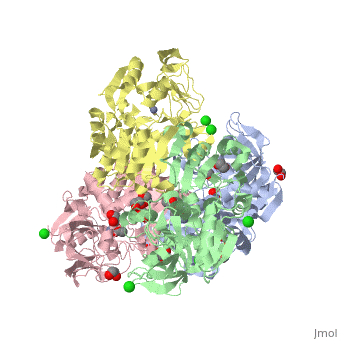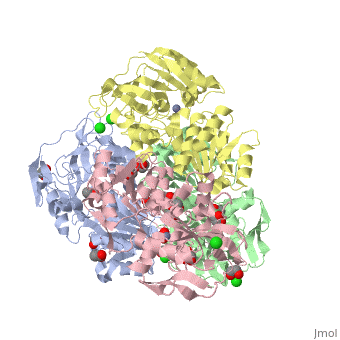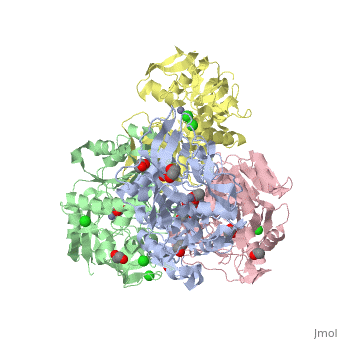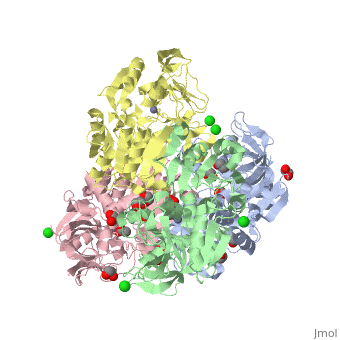
The double [http://en.wikipedia.org/wiki/Mutation mutant] of the chimera Χ21<sub>(TCT)</sub> (cofactor-binding domain of thermophilic TbADH replaced by that of mesophilic CbADH) Q165E/S254K-X21<sub>(TCT)</sub> ([[3ftn]]) was constructed by [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis]. In both TbADH and CbADH, Lys257 and Asp237 form an intrasubunit ion pair, in TbADH, Asp237 is also involved in an ion pair bridge with Arg304 of the adjacent monomer. In addition, Arg304 forms intersubunit salt bridge with Glu165 of the first monomer. Therefore, a <scene name='3fsr/Al/2'>four-member ion pair network</scene> involving Lys257, Asp237, and Glu165 of one monomer and Arg304 of the adjacent one is present in TbADH (the names of monomers are in brackets). However in mesophilic CbADH (and, therefore, in the chimera Χ21<sub>(TCT)</sub>, [[3fsr]]) the Gln is situated in position 165 (instead Glu of TbADH) and Met in position 304 (instead Arg of TbADH), so, such an ion pair network does not exist. In the double mutant Q165E/S254K-X21<sub>(TCT)</sub> reverse mutation Q165E reconstructs this network (as in parent thermophilic TbADH) that led to significant enhancement of the thermal stability of CbADH (ΔT<sub>1/2</sub><sup>60 min</sup> = 5.4 °C). <font color='magenta'><b>Chimera X21<sub>(TCT)</sub> ([[3fsr]]) is colored magenta</b></font> and <font color='cyan'><b>the double mutant Q165E/S254K-X21<sub>(TCT)</sub> cyan</b></font> ([[3ftn]]). In chimera X21<sub>(TCT)</sub>, position 254 is occupied by Ser (due to sequence of exchanged domain). The replacement of Ser254 of CbADH with Lys significantly enhances the stability of the enzyme, due to the formation of <scene name='3fsr/Al/3'>intrasubunit Lys254 and Glu280 ion pair</scene>. However, this replacing of Ser254 by Lys had a negligible effect on the thermal stability, in contrast to mutation Q165E mentioned above. | The double [http://en.wikipedia.org/wiki/Mutation mutant] of the chimera Χ21<sub>(TCT)</sub> (cofactor-binding domain of thermophilic TbADH replaced by that of mesophilic CbADH) Q165E/S254K-X21<sub>(TCT)</sub> ([[3ftn]]) was constructed by [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis]. In both TbADH and CbADH, Lys257 and Asp237 form an intrasubunit ion pair, in TbADH, Asp237 is also involved in an ion pair bridge with Arg304 of the adjacent monomer. In addition, Arg304 forms intersubunit salt bridge with Glu165 of the first monomer. Therefore, a <scene name='3fsr/Al/2'>four-member ion pair network</scene> involving Lys257, Asp237, and Glu165 of one monomer and Arg304 of the adjacent one is present in TbADH (the names of monomers are in brackets). However in mesophilic CbADH (and, therefore, in the chimera Χ21<sub>(TCT)</sub>, [[3fsr]]) the Gln is situated in position 165 (instead Glu of TbADH) and Met in position 304 (instead Arg of TbADH), so, such an ion pair network does not exist. In the double mutant Q165E/S254K-X21<sub>(TCT)</sub> reverse mutation Q165E reconstructs this network (as in parent thermophilic TbADH) that led to significant enhancement of the thermal stability of CbADH (ΔT<sub>1/2</sub><sup>60 min</sup> = 5.4 °C). <font color='magenta'><b>Chimera X21<sub>(TCT)</sub> ([[3fsr]]) is colored magenta</b></font> and <font color='cyan'><b>the double mutant Q165E/S254K-X21<sub>(TCT)</sub> cyan</b></font> ([[3ftn]]). In chimera X21<sub>(TCT)</sub>, position 254 is occupied by Ser (due to sequence of exchanged domain). The replacement of Ser254 of CbADH with Lys significantly enhances the stability of the enzyme, due to the formation of <scene name='3fsr/Al/3'>intrasubunit Lys254 and Glu280 ion pair</scene>. However, this replacing of Ser254 by Lys had a negligible effect on the thermal stability, in contrast to mutation Q165E mentioned above. | ||
| - | {{Clear}} | ||
| - | |||
| - | |||
| - | The <scene name='3fsr/Al1/2'>comparison</scene> of overall Cα backbone of all these chimeras (rmsd 0.45-0.65 Å) with those of the parent enzymes, did not reveal significant structural changes. So, the differences in the thermal stability of the chimeras and the parent enzymes could be caused by relatively small specific changes located at the important points of the NADP<sup>+</sup>-dependent alcohol dehydrogenases. For example see Cα superposition for the <font color='red'><b>X23<sub>(TET)</sub> chimera (red)</b></font> ([[3fpc]]) and its parent ADHs (<font color='blue'><b>TbADH, colored blue</b></font> ([[1ped]]), and <font color='lime'><b>EhADH1, colored lime</b></font> ([[1y9a]]). The [http://en.wikipedia.org/wiki/Root_mean_square_deviation RMSDs] of the TbADH−EhADH1, TbADH−Χ23<sub>(TET)</sub>, and EhADH1−Χ23<sub>(TET)</sub> were 0.68, 0.56, and 0.48 Å, respectively. | ||
| - | |||
| - | {{Clear}} | ||
</StructureSection> | </StructureSection> | ||
| Line 29: | Line 23: | ||
Biochemical and structural properties of chimeras constructed by exchange of cofactor binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms., Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon LJ, Frolow F, Burstein Y.Biochemistry. 2010 Jan 26. [http://pubs.acs.org/doi/abs/10.1021/bi901730x Epub ahead of print] | Biochemical and structural properties of chimeras constructed by exchange of cofactor binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms., Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon LJ, Frolow F, Burstein Y.Biochemistry. 2010 Jan 26. [http://pubs.acs.org/doi/abs/10.1021/bi901730x Epub ahead of print] | ||
* '''(See also [[Tetrameric alcohol dehydrogenases]])''' | * '''(See also [[Tetrameric alcohol dehydrogenases]])''' | ||
| + | * '''(See also [[Chimeres of alcohol dehydrogenases]])''' | ||
[[Category: Thermoanaerobacter brockii, entamoeba histolytica]] | [[Category: Thermoanaerobacter brockii, entamoeba histolytica]] | ||
[[Category: Burstein, Y.]] | [[Category: Burstein, Y.]] | ||
Revision as of 11:10, 12 June 2012
| |||||||||||
Reference
Biochemical and structural properties of chimeras constructed by exchange of cofactor binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms., Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon LJ, Frolow F, Burstein Y.Biochemistry. 2010 Jan 26. Epub ahead of print
- (See also Tetrameric alcohol dehydrogenases)
- (See also Chimeres of alcohol dehydrogenases)
Categories: Thermoanaerobacter brockii, entamoeba histolytica | Burstein, Y. | Felix, F. | Goihberg, E. | Shimon, L. | Bacterial alcohol dehydrogenase | Chimera | Cytoplasm | Domain exchange | Metal-binding | Nadp | Oxidoreductase | Zinc | ISPC, Israel Structural Proteomics Center. | Peretz, M. | Tel-Or, S. | ISPC | Israel Structural Proteomics Center | Structural genomic