Biological Unit: Showing
From Proteopedia
This article explains how to show a biological unit (biological assembly) from a green link in a scene in Proteopedia. If you simply want to see a biological unit, go to Visualizing the Biological Unit. For background, see Biological Unit and Asymmetric Unit.
Contents |
Biological Unit 1
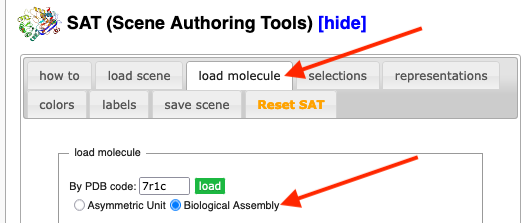
Proteopedia, using JSmol, is able to construct biological units. When you are developing a scene for a Proteopedia green link, using the Scene Authoring Tools with a published PDB code, you have the option of loading either the asymmetric unit or biological unit 1 (the "biological assembly").
Biological Unit 2 or Higher
Occasional PDB files specify more than one biological unit. The easiest way to see all the biological units is to use FirstGlance in Jmol: see Visualizing the Biological Unit.
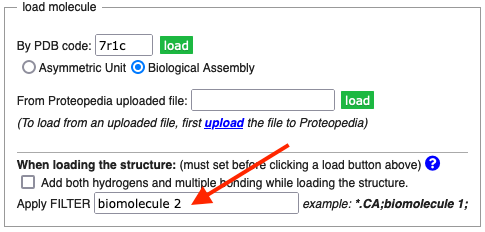
If you want to show biological unit 2 (or a higher number) in a green link scene in Proteopedia, use the FILTER option in the Load Molecule dialog of the Scene Authoring Tools. Biological units are designated as "biomolecules" in PDB files. To get biomolecule 2, simply enter biomolecule 2 in the FILTER slot. This will load all atoms in biomolecule 2.
Simplification
Sometimes there are too many atoms for good performance of JSmol, and in such cases, simplifying to show only the alpha carbons (or sometimes a subset of alpha carbons) is usually sufficient. Consider the biological units ("biomolecules" in PDB nomenclature) in 7r1c, a bacterial gas vesicle. Its asymmetric unit contains a single protein chain with 65 amino acids (alpha carbon atoms) and 496 atoms (hydrogen atoms were omitted).
| Biomolecule | Chains | Alpha Carbons | All Non-Hydrogen Atoms |
|---|---|---|---|
| 1 | 5 | 325 | 2,480 |
| 2 | 100 | 6,500 | 49,600 |
| 3 | 930 | 60,450 | 461,280 |
The performance of JSmol becomes sluggish with >30,000 atoms, and very sluggish with >100,000 atoms. FirstGlance simplifies the 930-chain biomolecule 3 of the 7r1c gas vesicle to every 3rd alpha carbon. In order to show the simplified model as a solid object, the "spacefilling" radii of the alpha carbons is expanded to 6.0 Å. The radius assigned to the alpha carbon atoms in models simplified by FirstGlance is reported when you click Solid in the Views tab.
Alpha Carbons Only
In order to load only the alpha carbon atoms, begin the FILTER with *.CA;. So, for example, the FILTER for biological unit 2 of 7r1c would be *.CA;biomolecule 2;. It is important to put a semicolon after each part of the FILTER.
Subsets of Alpha Carbons
When the number of alpha carbons exceeds about 30,000, JSmol will perform much better if only a subset of the alpha carbon atoms are loaded. To load only every 3rd alpha carbon, put /=3 at the end of the FILTER. So the complete FILTER could be, for example, *.CA;biomolecule 3;/=3;.
Load Filters
The filter used by FirstGlance, and in fact the entire load command, can be displayed by clicking Troubleshooting below the molecule. Then, scroll down to the bottom in the Troubleshooting (lower left) panel. Here is the report for 7r1c:
The Jmol command that loaded this file was:
load MODELS ({0}) https://files.rcsb.org/download/7R1C.pdb filter
"!_H;*.CA;*.P;%A;ALLHET;biomolecule 3;bmchains;/=3"; delete water;delete hydrogen;
Here is an explanation:
- MODELS ({0}) -- When multiple models are present in the PDB file, only the first model is loaded, regardless of whether that model is assigned number 1 (typical) or another number, such as number 0 (AlphaFold). When the PDB file contains only a single model, this has no effect.
- !_H; Do not load hydrogen atoms.
- *.CA; Load only alpha carbon atoms for proteins.
- *.P; Load only backbone phosphorus atoms for nucleic acids.
- %A; When some atoms have alternate locations, load only their first location. This avoids having multiple copies of the same atom in different locations.
- ALLHET; Load all hetero atoms (ligands). In addition to ligands, this loads the hydrogen atoms that are part of ligands or other hetero moieties, and it loads water.
- biomolecule 3; This constructs and loads biological unit (assembly) number 3 specified in the PDB file.
- bmchains; When some chains are duplicated to construct the biomolecule, each chain is given a distinct name. For example, 3hyd contains a singe chain A in its asymmetric unit, and 4 chains in biomolecule 1. The additional chains are named A2, A3, and A4. These are not legal chain names in PDB format, which restricts chain names to a single character. However, they work internally within JSmol and FirstGlance is adapted to handle them correctly. To see the complete list of chain names, click on the count of chains in the Molecule Information Tab of FirstGlance.
- /=3; This loads only every third alpha carbon or nucleic backbone phosphorus atom.
- delete water; This deletes all water that was loaded due to ALLHET.
- delete hydrogen; This deletes ligand hydrogen atoms that were loaded due to ALLHET.