Publication Abstract from PubMed
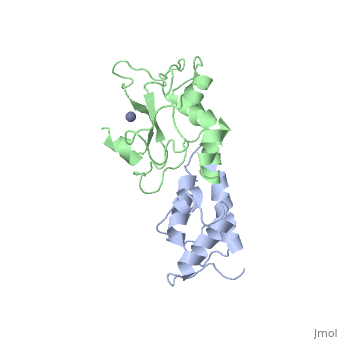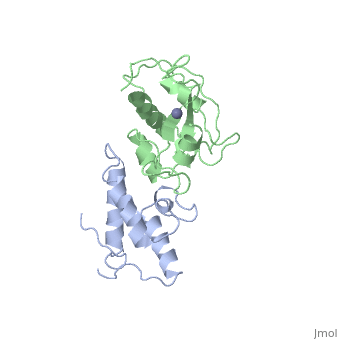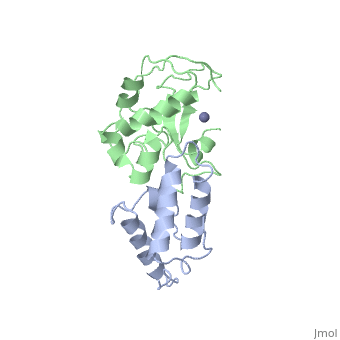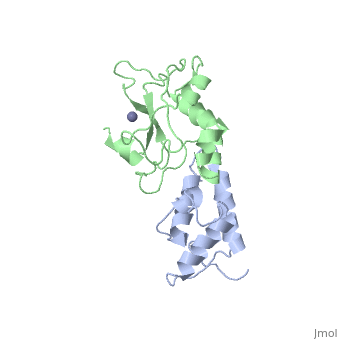
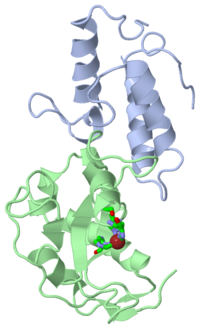
BACKGROUND: Colicin E7 (ColE7) is one of the bacterial toxins classified as a DNase-type E-group colicin. The cytotoxic activity of a colicin in a colicin-producing cell can be counteracted by binding of the colicin to a highly specific immunity protein. This biological event is a good model system for the investigation of protein recognition. RESULTS: The crystal structure of a one-to-one complex between the DNase domain of colicin E7 and its cognate immunity protein Im7 has been determined at 2.3 A resolution. Im7 in the complex is a varied four-helix bundle that is identical to the structure previously determined for uncomplexed Im7. The structure of the DNase domain of ColE7 displays a novel alpha/beta fold and contains a Zn2+ ion bound to three histidine residues and one water molecule in a distorted tetrahedron geometry. Im7 has a V-shaped structure, extending two arms to clamp the DNase domain of ColE7. One arm (alpha1(*)-loop12-alpha2(*); where * represents helices in Im7) is located in the region that displays the greatest sequence variation among members of the immunity proteins in the same subfamily. This arm mainly uses acidic sidechains to interact with the basic sidechains in the DNase domain of ColE7. The other arm (loop 23-alpha3(*)-loop 34) is more conserved and it interacts not only with the sidechain but also with the mainchain atoms of the DNase domain of ColE7. CONCLUSIONS: The protein interfaces between the DNase domain of ColE7 and Im7 are charge-complementary and charge interactions contribute significantly to the tight and specific binding between the two proteins. The more variable arm in Im7 dominates the binding specificity of the immunity protein to its cognate colicin. Biological and structural data suggest that the DNase active site for ColE7 is probably near the metal-binding site.
The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein., Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS, Structure. 1999 Jan 15;7(1):91-102. PMID:10368275
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
of the immunity protein 9 (Im9, 1bxi, colored yellow), evolved variant R12-2 (lime), and immunity protein 7 (Im7, colored blue, 7cei) reveals their structural identity. However, when the immunity proteins-bound , they demonstrate somewhat different picture. The Im9 and Im7 are differ more in their binding configurations (19°, with Tyr54-Tyr55 as the pivot), while the variant R12-2 is in an intermediate configuration between Im9 and Im7. Of note, in the variant R12-2 (3gkl) and Im9 (1bxi) there are Tyr54 and Tyr55, while in the Im7 (7cei) Tyr55 and Tyr56 are homologous to them. The most are in the loop between helices α1 and α2 in Im9 (yellow, labeled in black) and evolved variant R12-2 (lime, labeled in black). This loop consists of three mutations: N24D, T27A, and S28T in variant R12-2. We can see the deviations in the relative position of helices α1 and α2, in the loop's backbone and in the side chains of residues 24, 26 and 28.
Comparison of the different Im-colicin complexes reveals changes in the binding configuration of the evolved variants which increase affinity toward ColE7 by re-aligning pre-existing Im9 residues. Glu30 of Im9 (1bxi, colored yellow) forms with Arg54 of ColE9 (orange), whereas Asp51 have not direct side chain–side chain interactions. Asp31 of Im7 (blue) which is corresponding to Im9 Glu30 is involved in to Arg520 and Lys525 of ColE7 (darkmagenta), while Asp52 of Im7 (corresponding to Im9 Asp51) is within hydrogen bond distance to Thr531 and Arg530 of ColE7 (7cei). Glu30 in the variant R12-2 (lime) is shifted and forms a to Arg520 of ColE7 (magenta). Asp51 is within hydrogen bond distance to Thr531 of ColE7 (3gkl). However, the side chains of Lys525 and Arg530, which are very important in salt bridge contacts with Glu30 and Asp51, respectively, in the structure of the ColE7–Im7 complex have a different conformation that eliminates these contacts in evolved variant R12-2.