Function
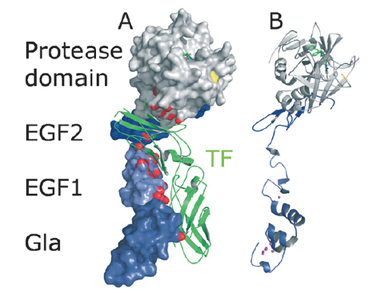
Factor VIIa (FVIIa) is a single chain trypsin-like serine protease [1](EC 3.4.21.21) of 406 residues. The FVII[2] zymogen is a glycoprotein consisting of an amino-terminal (N-linked) γ-carboxyglutamic acid (Gla)[3]domain followed by two epidermal growth factor-like (EGF1 and EGF2) domains, a short linker peptide, and a carboxy terminal serine protease domain[1]. The active form, FVIIa, is generated by a specific cleavage of a peptide bond between Arg152 and Ile153 at the end of the linker peptide by either factor Xa (FXa) or thrombin (IIa). This cleavage generates an N-terminal light chain of 152 residues linked to a heavy chain of 254 residues by a disulfide bridge [2]. Following cleavage the newly formed N-terminal inserts itself into a cavity, or the activation pocket, forming a salt bridge with Asp343 (Asp194 trypsin numbering).Formation of this salt bridge allows for the maturation of FVIIa to its active form. The images correspond to one representative Factor VIIa, i.e. the crystal structure of Human factor VIIa complex with tissue factor and a peptide inhibitor (1dan).
complex with .
.
.
(1dan).[3] Water molecules are shown as red spheres.
FVIIa mechanism
General
FVIIa alone shows very little proteolytic activity and only becomes fully active when complexed to its obligatory cofactor, tissue factor (TF) and cations, mainly Ca++. TF, located in the vessel wall, is exposed to circulating FVIIa upon injury or some type of stimulus and forms a TF-FVIIa complex. A unique property of TF-FVIIa among other coagulation enzyme complexes is that phospholipids are not an obligate requirement for the assembly of the complex. However, the activity of the complex towards its substrates (FIX and FX) requires a lipid surface which is provided by the membrane-anchored TF. The TF-phospholipid complex enhances the efficiency (kcat/Km) of FVIIa-catalyzed reactions by the 10^10-fold. There are four distinct steps that are required for the full activity of the TF-FVIIa complex: 1) proteolytic activation of single-chained FVII to two-chain disulfide bridged FVIIa 2) binding of cations 3) interaction of TF with FVIIa 4) acidic-membrane association and proper orientation of substrate[4][5].
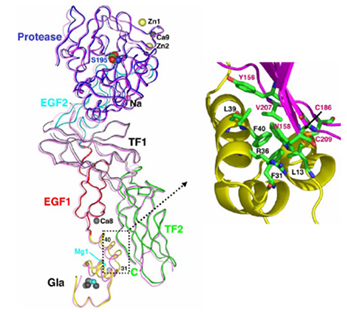
See also Mg-8 may contribute to the binding to factors VIIa and X.
Allosteric activation
Cation interaction
The Gla domain binds four Ca++ ions and three Mg++ ions arranged in a linear fashion. Ca++ induced changes in the Gla domain are responsible for major structural rearrangements in a region that facilitates binding of FVIIa to the membrane[6]. Binding of Ca++ induces an increase in the α-helical content of that region. The EGF1 domain contains one Ca++ binding site believed to be important for TF binding. The protease domain binds one Ca++, one Na+ and two Zn++ ions. The Zn++ ions have been shown to inhibit the activity of FVIIa specifically by reducing its affinity for TF. Ca++ binding in the protease domain, mediated by Glu210 and Glu220, produces subtle local changes presumably important for TF binding. The Na+ binding site is located in a hydrophobic cavity responsible for TF binding[7].
TF interaction - entropy trap
The binding epitope of TF to FVIIa is a stripe running along the whole length of the TF protein.
The Gla domain of FVIIa binds to the C-domain of TF. The interaction is mainly hydrophobic termed the “hydrophobic stack”. EGF1 domain packs into a groove formed by the two modules of TF. This interface is the largest and contributes the most energetically in binding of the cofactor to the protease domain. EGF2 and the catalytic domain interact with the N-domain of TF. Two “lock and key” interactions are observed. One is the side chain of Phe50 of TF which is trapped in the pocket formed by the end of EGF2 domain of FVIIa. Second, the side chain Met306 of FVIIa is enclosed by TF residues Arg74, Phe76, Glu92, Leu94. This Met306 residue is responsible for the thermodynamic coupling to the active site and is unique to FVIIa. Mutation of this residue results in the failure of TF to decrease the dissociation rate of the enzyme from the cofactor. Cofactor interactions, specifically through Met306 lead to subtle changes which then influence the position of Asp331. Asp331 is the specificity-determining residue in the binding pocket. Constraint and stabilization promote formation of a hydrogen bond between the amide of Arg315(170C) and carbonyl of Gly372(223). When bound to TF the activation region of FVIIa contains a large number of hydrogen bonds between the main chain and side chain atoms. At least 11 water molecules are identified in the catalytic domain region. An interesting observation is that the center of this region contains water, in contrast to the hydrophobic interactions in the other two interface regions. These hydrophilic interactions may be more efficient at mediating TF affinity and substantial conformational changes to induce activity of FVIIa. Therefore TF and cations are obligatory cofactors in the allosteric regulation of FVIIa activity by stabilizing the disordered, flexible FVIIa and restraining the enzyme for catalytic activity.
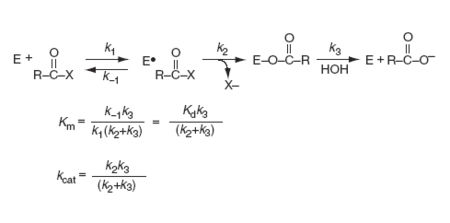
Kinetics of action
In general serine proteases have a three-step kinetic mechanim: 1) formation of an enzyme-substrate (E-S) complex(k+1, k-1), 2) acylation of the active site serine(k2), and 3) hydrolysis of the acylenzyme intermediate (k3).
Catalytic domain
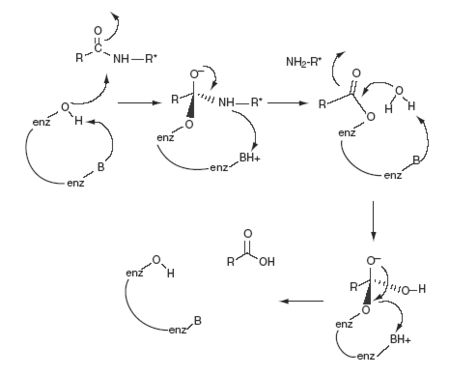
There are three steps serine proteases take to hydrolyze an amide bond: 1)activation of amide bonds by the interaction of the general acid with the carbonyl oxygen of the substrtate which disrupts resonance stabilization 2) activation of water by general base 3)activation of amines by protonation before expulsion. Serine proteases hydrolyze amide bonds with rates of 10^10–fold higher than the uncatalyzed reactions.
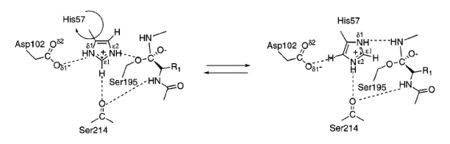
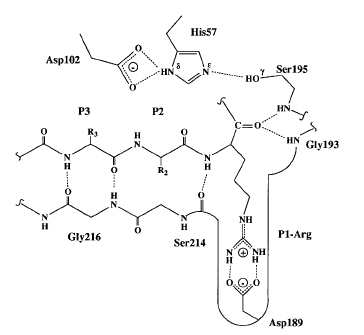
In FVIIa Ser344(195) of the catalytic triad is activated by a His193(57), or the general base, which itself is stabilized by a hydrogen bond to Asp242(102). These reactions result in a formation of a tetrahedral intermediate and the oxyanion hole. The oxyanion hole is stabilized by interactions with main chain NHs. The activated Ser344(195) then attacks the scissile bond of the substrate. The general base His193(57) transfers the abstracted proton from Ser to the amine leaving group, the tetrahedral intermediate (transition state) collapses and an acylenzyme intermediate is formed releasing the product. The general base His193(57) abstracts a proton from water as it attacks the acylenzyme to again form a tetrahedral intermediate. His193(57) then acts as an acid and protonates Ser344(195) releasing the product acid and regenerates the enzyme. This reaction is largely possible by having a His193(57) with a pKa ~7 necessary for deprotonation, a hydrogen bonding network or “the charge relay system” activating Ser344(195) for nucleophilic attack, stabilization of the negatively charged oxyanion of the tetrahedral intermediate by the main chain NHs of Ser344(195) and Gly342(193).
One-proton versus Two-proton transfer
The charge relay system explains the action of Ser344 as part of resonance forms Asp-CO2-/H-His/Ser-OH and Asp-CO2H/His-H/Ser-O which involve proton transfers between Ser344 to His193 and His193 to Asp242. However this two-proton transfer mechanim requires that the pKa of His344 is lower than pKa of Asp242. The argument for one-proton transfer mechanism, supported by 1H NMR experiments, observed the pKa of free enzyme and acylenzyme His193 to be ~7 whereas that of tetrahedral intermediate to be more than 10. This argues for electrostatic stabilization of protonated His193 by the negatively charged Asp242 or a one-proton transfer mechanims[8].
Low barrier hydrogen bonds (LBHB)
The hydrogen bond between Asp242 and His193 has been suggested to fulfill many criteria for low barrier hydrogen bond. This criteria includes a <2.6Ǻ distance between Asp242 and His193,an unusually downfield signal of the Nδ1 proton of His193 in 1H NMR, an observed deuterium isotope effect, and a low D/H fractionation factor of the Nδ1 proton[9].The arguments in opposition to LBHB include unmatched pKas of His193 and Asp242, localization of the proton mainly on His193 as determined by 15N-H spin couplings and 15N chemical shifts, lack of sheilding of the Nδ1 proton from water and polar environment, and multiple hydrogen interactions of both Asp242 and His193 which would weaken the Oδ1-Nδ1-H hydrogen bond[10].
"His Flip" mechanism
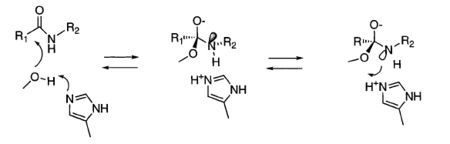
This mechanism argues that the protonation of the substrate leaving group by His193 is equally favorable as re-protonation of Ser344 and regeneration of substrate. In order to prevent regeneration of the substrate, His193-H+ flips placing the Nδ1 proton near the leaving group. The argument against this mechanims is the need of disruption and reformation of many hydrogen bonds which seems unlikely in the short lifetime of the transition state. Additionally the principle of loss of motion would be violated[11][12].
Stereochemistry of the tetrahedral intermediate
Upon attack on an amide bond, the lone electron pairs of the oxyanion and the nitrogen of the leaving group have to be antiperiplanar to the new bond. Therefore the lone pair of the amine leaving group points away from His193-H+. For the chemistry to occur the nitrogen must undergo inversion to position the lone pair for protonation by His193-H+[13].
Substrate binding induces the formation of the oxyanion hole
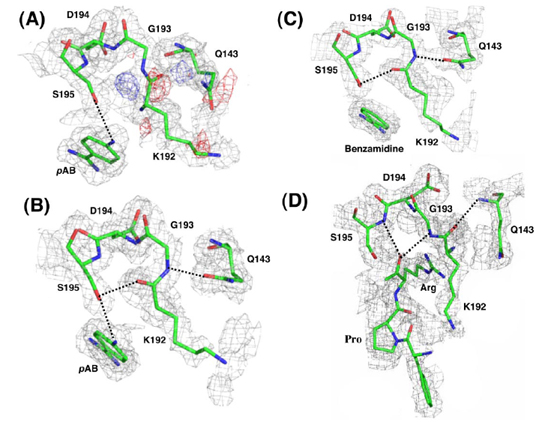
Atypical conformation of the Lys192-Gly193 peptide bond has been observed. The carbonyl O of Lys192 which normally points away from the oxyanion hole makes a H-bond with the gamma O of Ser195 while the side chain of Glu143 fomrs a H-bond with the amide of Gly193. This Lys192-Gly193 peptide bond conformation in FVIIa is unconventional. This bond flips to the proper orientation only upon substrate binding. This suggest that it is the substrate binding and not the TF cofactor binding that induces the oxyanion hole formation and functional active site geometry[14].
Evolutionary specificity
All of the serine proteases have a catalytic domain consisting of two β-barrels with the catalytic triad, Ser-His-Asp located at the interface[15]. Five enzyme-substrate hydrogen bonds at positions P1 and P3 are well conserved and serve to position the scissile peptide bond in the correct orientation for an attack by the γ-oxygen of Ser. More distal contacts diverge. Mutational experiments have shown that the S1 pocket is important in organization of the substrate for catalysis. Divergent evolution[4]enabled mammals to possess multiple enzymes with specific roles. Earlier organisms, bacteria and prokaryotes, possess broad specificities of the active site where more distant residues play a bigger role in substrtate recognition[16]. The most divergent are the surface loops which control specificity. Serine proteases not only show the divergence of substrate specificity but also examples of convergent evolution[5]. A demonstration of convergent evolution in serine proteases is found in four other folds, besides the chynotrypsin-like fold, with the catalytic triad in similar positions[17].
3D Structures of Factor VIIa
Factor VIIa 3D structures