Journal:JBIC:16
From Proteopedia

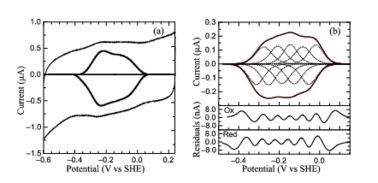
Laue Crystal Structure of Shewanella oneidensis Cytochrome c Nitrite Reductase from a High-yield Expression SystemMatthew Youngblut, Evan T. Judd, Vukica Srajer, Bilal Sayyed, Tyler Goelzer, Sean J. Elliott, Marius Schmidt and A. Andrew Pacheco [1] Molecular Tour Cytochrome c nitrite reductase (ccNIR) is a central enzyme of the nitrogen cycle. It binds nitrite, and reduces it by transferring 6 electrons to form ammonia. This ammonia can then be utilized to synthesize nitrogen containing molecules such as amino acids or nucleic acids. However, ccNiR’s primary role is to help extract energy from the reduction; ammonia is simply a potentially useful byproduct. In general, heterotrophic organisms feed on electron-rich substances such as sugars or fatty acids. During the metabolism of these substances large numbers of electrons are produced. Many organisms use oxygen as the final acceptor of these electrons, in which case water is formed. However, some organisms can use alternative electron acceptors such as nitrite, which is where ccNiR comes in. The ccNiR described here is produced by the Shewanella oneidensis bacterium, which is remarkable in its own right due to the large number of electron acceptors that it can utilize. Shewanella is a facultative anaerobe, which means that it will use oxygen if available, but in the absence of oxygen can get rid of its electrons by dumping them on a wide range of alternate acceptors, of which nitrite is only one example. To handle the electron flow Shewanella uses a large number of promiscuous containing electron transfer proteins. Indeed, Shewanella is exceptionally adept at producing c-heme proteins under fast-growth conditions, which many bacteria commonly used for large-scale laboratory gene expression, such as E. coli, are incapable of unless they are first extensively reprogrammed genetically. Since Shewanella can be easily grown in the lab, and can naturally and easily produce c-hemes, it is an ideal host for generating large quantities of c-heme proteins such as ccNiR. The 2012 paper by Youngblut et al. [1] describes a genetically modified Shewanella strain that can produce 20 – 40 times more ccNiR per liter of culture than the wild type bacterium. The ccNir so produced can be purified easily and in large amounts. This result is important because c-heme proteins have historically proved difficult to over-express in traditional vectors such as E. coli. With large quantities of Shewanella ccNIR available, Youngblut et al [1] were able to obtain the crystal structure (3ubr) and do a variety of experiments. The ccNIR consists of (colored in darkmagenta and in green) with . In the oxidized ccNIR all central heme irons are Fe3+. They can be subsequently reduced to Fe2+ either by reducing agents or electrochemically. An important conclusion of the paper is that electrons added to ccNiR are likely , rather than localized on individual hemes. The hemes 3-5 (colored in yellow) and the hemes-2 (colored in seagreen) are six coordinate and used for electron transport only, whereas the two hemes-1 (colored in magenta) are the active sites. Electrons are believed to enter via the hemes-2, but can move between subunits. Though the physiological significance of this result is not yet known, it is possible that delocalizing the electrons keeps the active site redox-potential sufficiently high until enough electrons are accumulated that the reaction with nitrite can take place. That is, CcNIR acts like a capacitor that can store electrons until they are needed. The X-ray structure of the ccNIR reveals the architecture of this capacitor. To solve the structure a non-standard method, the Laue method, was used. This became necessary since attempts to collect a high resolution data set with monochromatic X-ray radiation were not successful. At room temperature the ccNIR crystals are susceptible to radiation damage. Freezing damaged the crystals because a suitable cryoprotectant could not be found. Single pulsed Laue crystallography with 100 ps highly intense polychromatic X-ray pulses provided a solution. A dataset was collected in a few minutes. The crystals were cooled slightly to 0 °C but not frozen. Crystal settings spanned a range of 180 °C and the crystals were orthorhombic. Therefore, a Laue dataset with very high multiplicity and good quality in terms of resolution and Rmerge could be collected. The structure of this ccNIR was then solved by molecular replacement using the E. coli ccNIR as a template. of the S. oneidensis hemes within one monomer with the corresponding E. coli hemes reveals significant similarity. S. oneidensis hemes 3-5, hemes-2, and hemes-1 are colored in yellow, seagreen, and magenta, respectively, whereas their corresponding E. coli hemes are in similar, but darker colors. The of S. oneidensis ccNiR also is similar to that of E. coli ccNiR, except in the region where the enzyme interacts with its physiological electron donor (CymA in the case of S. oneidensis ccNiR, NrfB in the case of the E. coli protein) near heme 2. Subunits of S. oneidensis ccNiR colored in darkmagenta and in green; subunits of E. coli ccNiR colored in hot pink and in deep-sky-blue. Protein film voltammetry (PFV) experiments performed on S. oneidensis ccNiR films in the absence of substrate produced a broad envelope of reversible signals that span approximately 450 mV. At high pH values the envelope appears as a single peak, whereas at pH values below 7 the envelope appears to be composed of two large overlapping peaks. At pH values below 6 and at 0 °C, the envelope of signal can be better resolved and more than two peaks can be observed. This resulting envelope of signal can be deconvoluted as the sum of five one-electron peaks, each corresponding to one of the five hemes in a ccNiR monomer (see image below). The Ca2+ ion within is coordinated in bidentate fashion by , and in monodentate fashion by the backbone carbonyls, and the side-chain carbonyl. In the S. oneidensis structure only is assigned to the Ca2+ ion in subunit B. In subunit A the difference electron density that represents this water molecule is very close to the noise level, and it is difficult to identify even one water molecule there. The come near to the open calcium coordination sites, but are not within bonding distance. Instead they interact with the water molecule that is weakly coordinated to the Ca2+ ion. The ccNiR calcium ions appear to play a vital role in organizing the (as was mentioned above hemes-1 are the active sites). PDB reference: Laue structure of Shewanella oneidensis cytochrome-c Nitrite Reductase, 3ubr.
| |||||||||||
- ↑ 1.0 1.1 1.2 Youngblut M, Judd ET, Srajer V, Sayyed B, Goelzer T, Elliott SJ, Schmidt M, Pacheco AA. Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. J Biol Inorg Chem. 2012 Mar 2. PMID:22382353 doi:10.1007/s00775-012-0885-0