Molecular modeling study for conformational changes of Sirtuin 2 due to substrate and inhibitor binding
Sugunadevi Sakkiah, Meganathan Chandrasekaran, Yuno Lee, Songmi Kim, Keun Woo Lee [1]
Molecular Tour
, belongs to the member of NAD+-dependent deacetylase family. Structural detail of sirtuin 2 (SIRT2) complexes will be most utilitarian to discover the drug which might have a beneficial effects on a variety of human diseases like cancer, diabetes etc. Unfortunately till now there is a lack of SIRT2-ligand complex structure details, hence molecular docking was carried out to dock the substrate (NAD and acetylated lysine) and inhibitor (sirtinol) in the NAD binding pocket. The suitable binding orientation of (protein is colored in yellow, NAD is in magenta) and (protein is colored in green, inhibitor is in salmon), in SIRT2 active site were subjected to 5 ns molecular dynamics simulations to adjust the docked ligand structure into the active site and to identify its and dynamic behavior in active site. The result of our study affords an idea about the 3D structural details of SIRT2 in presence of substrate and inhibitor and their orientation in active site, the . In addition, the simulation revealed that the displacement of F96 upon substrate and inhibitor binding induced an and . We believed that our SIRT2 modeling studies could be much helpful to gain the structural insight of SIRT2 and also this will be useful to design the receptor-based inhibitors.
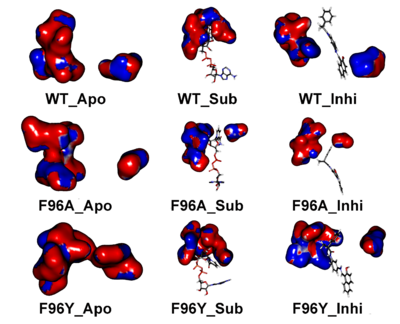
Electrostatic potential map indicates the assembly and disassembly of C pocket in the presence of NAD+ and inhibitor. Apo-form (left column), SIRT2–substrate (central column), and SIRT2–inhibitor (right column).


