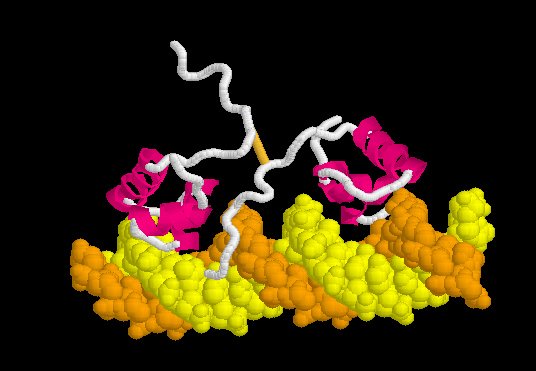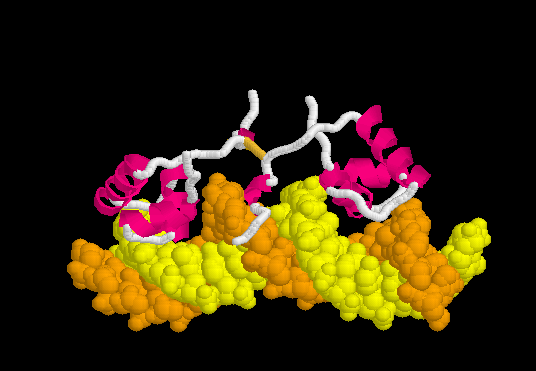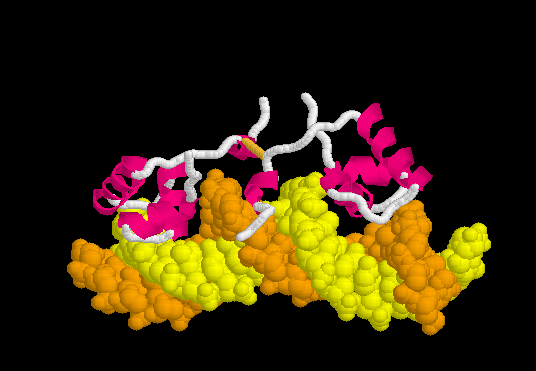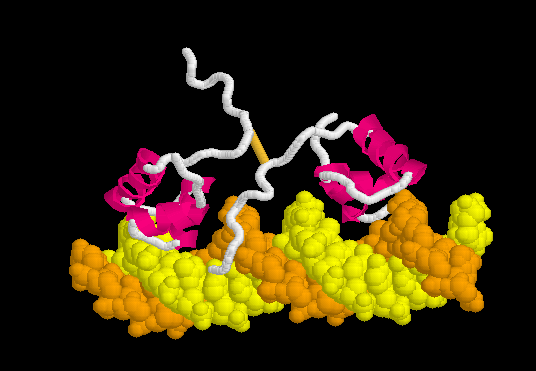
Morph of the lac repressor complexed with DNA
() After displaying interactive model:
showing the differences between non-specific binding (straight DNA) vs. specific recognition of the operator sequence (kinked DNA). Whether the binding kinks the DNA, or simply stabilizes a pre-existing kink, is unknown. Details Below.
What is the lac repressor?
Repressors are proteins that inhibit the expression of genes; that is, they inhibit the transcription of messenger RNA from their target genes. Each repressor targets a specific co-regulated group of genes by recognizing a specific sequence of DNA, called the operator in bacteria. Repressor proteins are coded for by regulatory genes.
The lactose ("lac") repressor controls the expression of bacterial enzymes involved in the metabolism of of the sugar lactose. When the lac repressor binds lactose, it changes to an inactive conformation that cannot repress the production of these enzymes. Thus, the enzymes needed to use lactose are made only when lactose is available. The lac repressor, and the group of genes it controls, which is called an operon, were the first such gene regulatory system to be discovered. The operon was described in 1960[1] by François Jacob et al., who also correctly proposed the general mechanism of regulation by the lac repressor. The 1965 Nobel Prize in Physiology or Medicine was awarded to François Jacob, André Lwoff, and Jacques Monod "for their discoveries concerning genetic control of enzyme and virus synthesis".
For a general introduction to the lac repressor, please see David Goodsell's Introduction to the lac repressor in his series Molecule of the Month, and the article in Wikipedia on the lac repressor. Mitchell Lewis published a detailed review in 2005[2]. See also Transcription and RNA Processing.
Structure of the lac repressor
The lac repressor protein ( showing chain A in 1lbg, resolution 4.8 Å), starting at the N-terminus, begins with a DNA-binding "headpiece", followed by a hinge region, then an N-terminal ligand-binding subdomain and a C-terminal ligand binding subdomain, a linker, and a C-terminal tetramerization helix[3]. (.) In the absence of DNA, the hinge region does not form the alpha helix shown here.
As can be seen when the chain is
each of the ligand-binding subdomains is made up of two discontinuous segments.
The lac repressor forms . Dimerization buries 2,200 Å2 of surface, including a ,
Hydrophobic, Polar
forming a hydrophobic core (shown with 1lbi, resolution 2.7 Å, lacking the DNA-binding domain due to disorder).
The most highly is the surface that contacts DNA[4]. (Only alpha carbon atoms are shown here, without sidechains, because sidechains were not resolved in the 4.8 Å 1lbg model.) The dimerization surfaces are the of the ligand-binding domains[5]. (This scene shows sidechains, using the 2.7 Å model in 1lbi, which lacks the DNA-binding domain due to disorder.)
The C-terminal tetramerization helices tether two dimers, and thus the functional form of with two DNA-binding sites.
DNA Binding: A Kink In The Operator
Non-Specific Binding
Lac repressor binds to DNA non-specifically ( derived [6] from 1osl, 20 NMR models), enabling it to slide rapidly along the DNA double helix until it encounters the lac operator sequence ("facilitated diffusion"[7]). The DNA-binding domain employs a helix-turn-helix motif (Alpha Helices, Turns). During non-specific binding, the hinge region is disordered (indicated by the range of positions of the 20 models). The DNA double helix is depicted as straight in the model shown here (see methods), but in actuality, straightness likely varies with sequence (see below). The protein model shown at right (1osl) has two copies of the DNA-binding domain and hinge region ( to distinguish the chain B hinge). these 20 NMR models simulates thermal motion of the disordered hinge regions.
Specific Binding
Upon recognizing the specific operator sequence, the non-specific binding converts to (derived[6] from 1l1m, 20 NMR models). During this conversion, the hinge region changes from disordered loops to Alpha Helices (: toggle spinning off to see highlighting), which bind to the minor groove of the DNA. As explained below, this binding stabilizes a kinked ("bent") DNA double helix conformation. What percentage of time this DNA sequence spends in a kinked state, in the absence of bound lac repressor protein, is not known, but it may be a significant percentage (see next section below). these 20 NMR models can be compared with the animation of the non-specific binding. See Lac repressor morph methods.
DNA Recognition
Proteins typically recognize specific DNA sequences in the major groove by direct readout, that is, by forming sequence-specific hydrogen bonds with the edges of the DNA bases exposed in the major groove[8].
DNA sequence recognition in the minor groove, often accompanied by kinking or bending of the DNA, is more complex. Direct readout is less important, since, unlike in the major groove, the four bases do not present unique hydrogen-bonding surfaces in the minor groove[8]. Recognition of the shape of the DNA seems more important[9][10]. In many cases, cationic arginines are believed to be attracted to a region of the minor groove with high aninoic charge density resulting from narrowing of the groove[10]. In these cases, the protein appears to recognize the shape of the DNA minor groove (indirect readout)[10].
In the lac repressor complex with specific DNA, a pair of arginines (Arg51 in each chain) is close to the minor groove, but points away from the groove (). Hence the binding of arginines to narrow minor grooves does not appear to be involved in specific DNA recognition by the lac repressor.
DNA Kinks
Strictly speaking, bends in DNA are distinguished from kinks. DNA is said to be kinked when the stacking contact between two adjacent base pairs is disrupted[8]. The DNA on either side of a kink may be straight or bent. A : a single CpG base pair is partially separated from the adjacent CpG base pair. . Pyrimidine-purine base pairs have the weakest stacking interactions, and are most susceptible to kinking[8]. In the complex of lac repressor with specific DNA, (if scene is blank,) are partially intercalated between the separated CpG base pairs, which helps to stabilize the kink. It may often be the case that sequence-dependent kinks and bends are present in DNA prior to the binding of protein[8]. DNA structure is dynamic. For example, recently Hoogsteen base pairing was observed to occur transiently in equilibrium with Watson-Crick base pairing[11] (See News & Views[12]). Also, the binding of p53 to some but not all DNA sequences stabilizes Hoogsteen (rather than Watson-Crick) base pairing[13]. Thus, the "bending" (actually kinking) depicted in the morph on this page may give the wrong impression: lac repressor binding may simply stabilize a kink (or transient kink) that pre-existed in the cognate DNA sequence.
DNA Bends
Strictly speaking, bends in DNA are distinguished from kinks. Bending means a curvature distributed over
several adjacent base pairs[8], whereas a kink (see previous section above) is a disruption in stacking limited to a single pair of adjacent base pairs. An is found in the DNA complexed to HPV E2 protein (thanks to [8] for this example, 1jj2).
Morph of Conversion
The can be seen more easily when they are animated smoothly by morphing. (The methods used to create this morph are given in Lac repressor morph methods.)
- Note the kinking of the DNA, with the widening of the central minor groove on the convex aspect. Whether the cognate DNA sequence is (perhaps transiently) kinked prior to lac repressor binding is unknown (see above).
- Also note the conversion of the disulfide-bonded hinge region loops to alpha helices. (The displayed secondary structure is calculated for each model in the morph interpolation.)
The specific recognition of the lac operator sequence in the DNA occurs largely though hydrogen bonds. is illustrated in this rendering of the morph. Shown are hydrogen bonds involving Arg22.N-eta2 and Tyr18.OH interacting with DNA base oxygens in the major groove, and Ala53.O interacting with a DNA base nitrogen in the minor groove. (Not all of the relevant hydrogen bonds are shown; see Methods.)
Animation for Powerpoint® Slides
Here is an animated multi-gif true movie of the above morph, ready to insert into a Powerpoint®[14] slide. If the image below is not moving, reload this page (it stops after 50 cycles).
- In Windows, simply drag the movie and drop it into the Powerpoint slide. You can then resize it and position it. The movie should play when you change the View to Slide Show ("project") the slide.
- In Mac OSX, Ctrl-Click on the movie, then Save Image. In Mac Powerpoint, at the desired slide, use the Insert menu (at the top) and select Movie ..., then insert the saved .gif movie file. After inserting the movie, make sure the Toolbox is showing (controlled with an icon-button at the top of the window). Now you can resize and reposition the movie. Click in the movie in the slide to select it. Now, in the Toolbox/Formatting Palette, under Movie, check Loop Until Stopped. Now the movie should play when you change the View to Slide Show ("project") the slide.
Challenge Your Understanding
Here are some questions to challenge your understanding.
- Why does the lac repressor bind to DNA non-specifically?
- When the lac repressor binds non-specifically to DNA, what part of the DNA double helix does it bind to?
- Does DNA have a net charge, and if so, is it negative or positive in aqueous solution at pH 7?
- What kinds of chemical bonds are likely to be involved in non-specific binding of the repressor protein to DNA?
- Does specific binding of lac repressor to DNA disrupt any of the Watson-Crick hydrogen bonds between the base pairs in the DNA strands?
- How do proteins such as the lac repressor recognize specific nucleotide sequences in a DNA double helix?
- What kinds of chemical bonds are involved in specific binding of the repressor protein to DNA?
- Does the lac repressor recognize specific bases in the major or minor grooves of the DNA?
- When it recognizes its specific nucleotide sequence, how does the lac repressor stabilize a kink in the DNA double helix?
Answers are available on request to  . If you would like us to make the answers publically available within Proteopedia, please let us know. When contacting us, please give your full name, your position, institution or school, and location.
. If you would like us to make the answers publically available within Proteopedia, please let us know. When contacting us, please give your full name, your position, institution or school, and location.
Content Attribution & Acknowledgement
The morphs displayed here were originally prepared by Eric Martz in 2004 for the page Lac Repressor Binding to DNA, within ProteinExplorer.Org.
Eric Martz thanks Remo Rohs for his kind and expert advice concerning the 2010-2011 updates to this article.