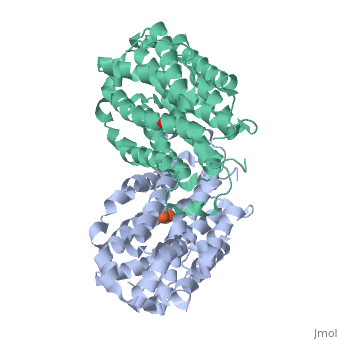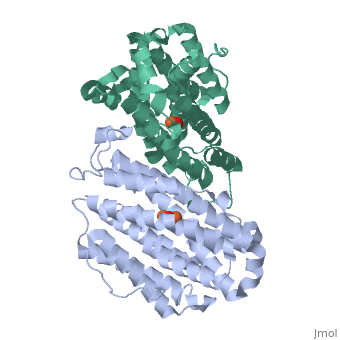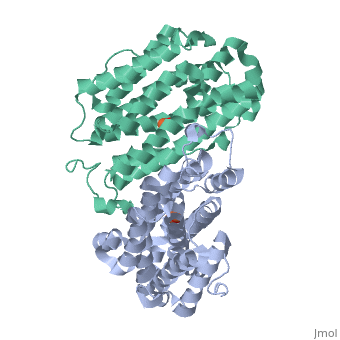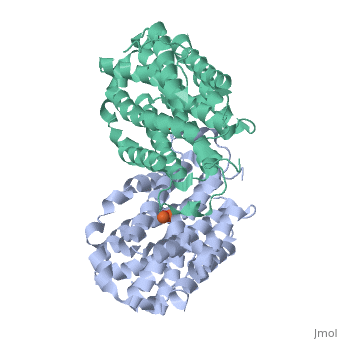
Mouse Ribonucleotide Reductase R2 (EC 1.17.4) is a class I enzyme. Class I enzymes consist of two different subunits. These two subunits are connected via hydrogen bonds. This enzyme acts primarily to catalyze the reduction of nucleoside diphosphates to deoxynucleoside diphosphates. Within DNA synthesis, this is the first dedicated step. The R2 subunit, a homodimer, is the smaller of the two subunits in this enzyme and serves as a limiting agent for overall enzyme activity. This subunit acts to maintain the tyrosil radical neighboring a diiron carboxylate site. In the mouse R2 subunit, the tyrosine residue is Tyr177. Through the process of catalysis, it has been proposed that the oxidation equivalent stored in the tyrosil subunit is passed along a pathway to a cysteine residue at an active site. Both subunits act as electron transfer pathways to the iron site. An arginine residue, Arg 265, has proven to be key to the activity of this particular enzyme. [1]
Activity Within DNA Synthesis
Ribonucleotide reductase is essential in the formation of new deoxyribonucleotides. These are essential for the repair and replication of DNA with organisms. This reaction is highly regulated, which is important; since levels of deoxyribonucleotides must be correct to lower chances of cell death or genetic abnormalities. With mice and humans, the two subunits are coded for by genes located on different chromosomes. The R2 subunit determines the activity of the enzyme, since its levels vary while the levels of the R1 subunit are quite constant. DNA damage induces transcription of the R1 gene only. The transcription of R2 is induced by p53 during the S phase of the cell. One interesting fact about this gene is that the human ribonucleotide reductase R2 promoter can activate transcription of the human R2 gene in a mouse cell. The human R2 gene can be expressed in a functional R2 protein within a mouse cell. The same is true for a mouse R2 gene in a human cell. This illustrates close relatedness of the genes. [2]
Structual Properties
While both forms of this enzyme both interact with iron, there are some structural differences due to the different conditions.
:
16 α-helices
8 β-turns
:
15 α-helices
10 β-turns[3]
Protein Sequence
For the enzyme under oxidizing conditions (1w68):
VEDEPLLRENPRRFVVFPIEYHDIWQMYKKAEASFWTAEEVDLSKDIQHWEALKPDERHFISHVLAFFA
ASDGIVNENLVERFSQEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYIKDPKEREYLFNAIETMPCVK
KKADWALRWIGDKEATYGERVVAFAAVEGIFFSGSFASIFWLKKRGLMPGLTFSNELISRDEGLHCDFA
CLMFKHLVHKPAEQRVREIITNAVRIEQEFLTEALPVKLIGMNCTLMKQYIEFVADRLMLELGFNKIFR
VENPF
For the enzyme under reducing conditions(1w69):
VEDEPLLRENPRRFVVFPIEYHDIWQMYKKAEASFWTAEEVDLSKDIQHWEALKPDERHFISHVLAFFA
ASDGIVNENLVERFSQEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYIKDPKEREYLFNAIETMPCVK
KKADWALRWIGDKEATYGERVVAFAAVEGIFFSGSFASIFWLKKRGLMPGLTFSNELISRDEGLHCDFA
CLMFKHLVHKPAEQRVREIITNAVRIEQEFLTEALPVKLIGMNCTLMKQYIEFVADRLMLELGFNKIFR
VENPF[4]