Function
H/ACA ribonucleoproteins that use RNAs to guide pseudouridine formation.
Beyond synthases
Interestingly, there are clearly-related complexes do not seem to guide pseudouridine formation. Similar complexes with the same proteins and similar RNAs are also known to be involved in cleavage of rRNA or stabilizing the ends of non-coding RNAs. Although they share even the same catalytic protein, the do not some to catalyze the reaction as with other substrates.
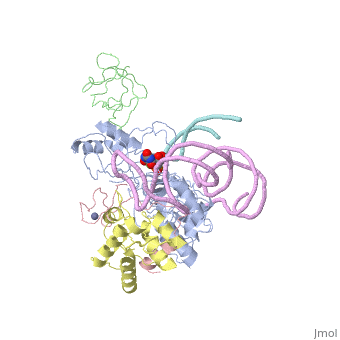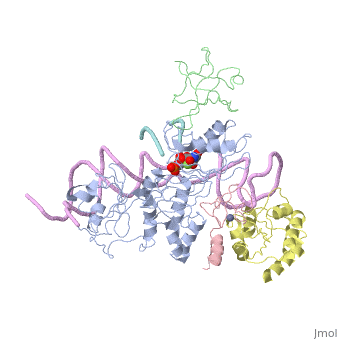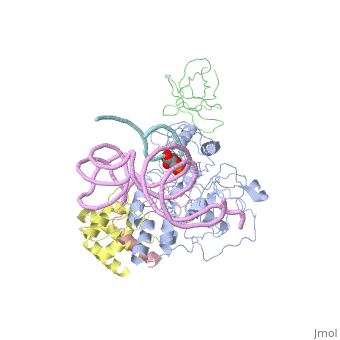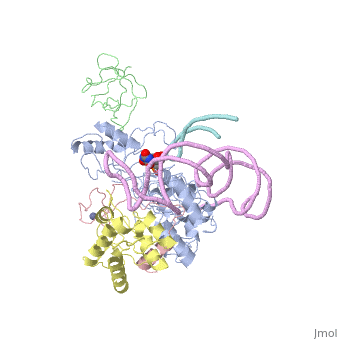
Structures of RNA-guided pseudoridine synthase
- Eukaryotic
- 3u28 Cbf5-Nop10-Gar1 complex (Saccharomyces cerevisiae)
- Archaeal
- 3hay - the full complex with substrate RNA (Pyrococcus furiosus)
- 3hjw - the Cbf5-L7Ae-Nop10-guide RNA-substrate RNA complex (Pyrococcus furiosus)
- 2hvy - Cbf5-Gar1-L7Ae-Nop10-guide RNA complex (Pyrococcus furiosus)
- 3hjy - the Cbf5-Nop10-guide RNA-substrate RNA complex (Pyrococcus furiosus)
- 2rfk - Cbf5-Gar1-Nop10-guide RNA-substrate RNA complex (Pyrococcus furiosus)
- 3hax - Cbf5-L7Ae-Nop10-guide RNA-substrate RNA complex (Pyrococcus furiosus)
- 2apo - Cbf5-Nop10 complex (Methanococcus jannaschii)
- 2aus - Cbf5-Nop10 complex (Pyrococcus abyssi)
- 3hay - the full complex with substrate RNA (Pyrococcus furiosus)
- 2ey4 - Cbf5-Gar1-Nop10 complex (Pyrococcus furiosus)