Student Project 1 for UMass Chemistry 423 Spring 2015
From Proteopedia
'Protein kinase C related kinase/Tofacitinib (prostrate and ovarian cancer)-4OTI'
by Emily Friis, Md Shafiul Hossain, Emily Hutchinson, Hieu (Vinny) La, John MacMunn, Kevin Purcell
Student Projects for UMass Chemistry 423 Spring 2015
Contents |
Introduction
|
Protein kinase C related kinase is a family of enzymes that is initiated by hydrolysis of inositol phospholipids. They transmit extracellular signals across membrane and helps regulate calcium ion dependent processes. helps elucidate a lot of biochemical mechanism of signal transduction and makes it easier for us to understand protein cell-to-cell communication. [1]
PRK1 is a component of Rho-GTPase, histone demethylase, androgen receptor, and histone demethylase signaling pathways and is involved in ovary and prostate cancer. A lot of PRK1 is expressed in cases of ovarian serous carcinoma. PRK1 is a family of AGC kinase enzymes. This group of enzymes has a C-terminal regulatory region. The C-tail helps regulate enzyme activity. The C-terminal can also recruit binding partners including PDK1. This phenylalanine region is the active-site tether and can connect to both nucleotide and inhibitors. [2]
Overall Structure
|
4OTI is a single chain protein, comprised of 15 alpha helices and 8 beta strands shown . The beta strands form , which are arranged antiparallel, giving added stability to the molecule, orienting its hydrogen bonds in a favorable manner. There are a total of 304 residues in the protein. [3]
4oti can be divided into , an N terminal lobe region providing regulation, and a C terminal lobe region that proposes as a catalytic region. The N and C terminals are separated by a hinge region, which can be adjusted when the enzyme is bound to a membrane. One key structural feature of 4oti is the C-tail, part of the , which travels from the C lobe to a final position in front of the N lobe. The lobe regions are approximately 20-40 kDa and 45kDa in size respectively. [4]
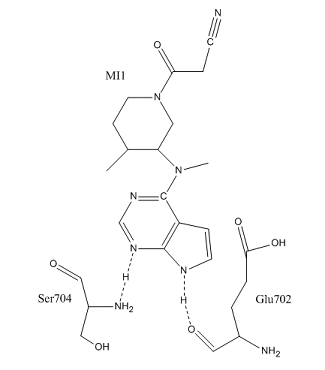
4oti's beta sheets reside in the N-lobe, and construct a , which serves as an open binding site for a ligand. is known as MI1, and has a structural formula of C16H20N6O. to the C-terminus of Glu702 and the N-terminus of Ser704, forming hydrogen bonds with Glu's carbonyl and Ser's nitrogen.
4oti's contains an additional 16 residues, which are available for only non-binding interactions. [3] Many of the of otherwise buried residues, such as Glu702 serve as binding sites for interactions with water.
Binding Interactions
|
Translocation into the plasma membrane is the ultimate goal of PRK1’s function and can only be achieved after proper activation by phosphorylation of the residues Ser 922, in the turn motif of N-lobe, and Thr 780 in the activation loop. The of residue Ser 922 includes a charges pocket of the residues Arg 629, Lys 634, and Lys 653, which further prompts the conformational changes in the “hinge point” between the C-terminal and N-terminal. This is required to activate substate and cofactor binding that leads to further conformational changes and eventual translocation and signaling.[5]
Cofactor binding, which is similar in all Protein Kinases, is located in the . In this case, the AST is centered around the residue Phe 910 that is stacked between Leu 627 and Gly 707, which directs the ATP coordination to the β3 strand in the C-helix. Another important area is the , which stabilizes the AST site and phosphorylation conformation changes, although it is less understood at this time[6].
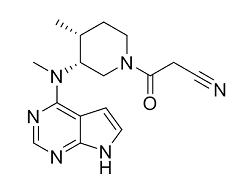
Most importantly, the AST region of the protein is the target area for protein inhibition in drug treatments. Known inhibitors such as Tofacitinib, lestaurtinib, and Ro-31-8220 interact with the binding site, specifically Phe 910, and disrupt ATP coordination as well as other secondary structure shifts. The MI1 ligand seen in the 4oti green scenes throughout this page is the bound Tofacitinib ligand.
Additional Features
|
In individuals with prostate and ovarian cancer, PRK1 is overexpressed in affected cells. Research has shown that higher expression levels correlate with Gleason scores in prostate cancer. Inhibition of PRK1 aids in preventing the proliferation of cancerous cells.[3]
PRK1 is capable of moving between the cytoplasm and nucleus. The activity levels of PRK1 can be controlled by a variety of small signaling molecules along with phosphorylation and proteolysis.
A characteristic feature of PRK1 is a C-terminal regulatory region. This section of the protein regulates enzyme activity and can recruit binding partner [6].The C-tail can be found at the base of the C-lobe and encircles the C-lobe. In addition to its main role in the cell, PRK1 also aids in the epigenetic regulation of transcription. A threonine residue, Thr 11 phosphorylates histone H3 and can enhance transcription of the nearby genes.[3]
PRK1 is inhibited by the naturally occurring molecule staurosporine. Clinical trials are being ran on several PRK1 inhibitors to observe their medical benefits. Even though PRK1 plays a role in pathways implicated by ovarian and prostate cancer, these inhibitors are being evaluated for treating other diseases. Lestaurtinib (CEP701) is a staurosporine analogue and inhibits several other protein kinases.CEP701 is being evaluated as a potential treatment for myelofibrosis and AML[5]. CEP701 causes a substantial disordering of the C-tail when it binds. Another drug, tofacitinib, has been approved for use as a rheumatoid arthritis treatment. Tofacitinib does not show the same displacement when it binds, but does make van der Waals Phe 910 . Instead, it results in a clashing with the sidechain of the G-loop residue and causes it to position away from the ATP binding site.
Quiz Question 1
|
4oti is a member of a family of Protein Kinase C related kinases, along with , , and .
Two inhibitors have been proposed for inhibiting PRK1s, Tofactenib:
And Lestaurtinib:
A) Which inhibitor is most likely to be successful on 4oti? Why? What type of inhibitor is it?
Consider the active site of and . Note the conservation of the Glu702 and Ser704 residues, and their positions compared to the beta sheets. Compare to , and notice that they both hydrogen bond to Glu702 and Ser704.
B) What does this tell you about the nature of the active site for , MI1, in 4oti compared to 4oth?
Quiz Question 2
|
a. Phe910
b. Pro 820
c. Arg 660
d. Both A and B
e. A, B, and C
See Also
Credits
Introduction - Md Shafiul Hossain
Overall Structure - Kevin Purcell & John MacMunn
Binding Interactions - Emily Hutchinson & Emily Friis
Additional Features - Hieu La
Quiz Question 1 - Md Shafiul Hossain, Kevin Purcell & John MacMunn , Emily Hutchinson & Emily Friis, Hieu La
Quiz Question 2 - Md Shafiul Hossain, Kevin Purcell & John MacMunn , Emily Hutchinson & Emily Friis, Hieu La
References
- ↑ "Studies and perspectives of protein kinase C." Science 233.4761 (1986): 305-312.
- ↑ Nishizuka, Yasutomi. <ref>DOI: 10.1371/journal.pone.0103638</li> <li id="cite_note-rasmol-2">↑ <sup>[[#cite_ref-rasmol_2-0|3.0]]</sup> <sup>[[#cite_ref-rasmol_2-1|3.1]]</sup> <sup>[[#cite_ref-rasmol_2-2|3.2]]</sup> <sup>[[#cite_ref-rasmol_2-3|3.3]]</sup> Chamberlain P, Delker S, Pagarigan B, Mahmoudi A, Jackson P, Abbasian M, Muir J, Raheja N, Cathers B. Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes. PLoS One. 2014 Aug 11;9(8):e103638. doi: 10.1371/journal.pone.0103638., eCollection 2014. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/25111382 25111382] doi:[http://dx.doi.org/10.1371/journal.pone.0103638 http://dx.doi.org/10.1371/journal.pone.0103638]</li> <li id="cite_note-3">[[#cite_ref-3|↑]] Newton, A. C. (1995). Protein kinase C: structure, function, and regulation. J. Biol. Chem. 270, 28495-28498. </li> <li id="cite_note-rasmo-4">↑ <sup>[[#cite_ref-rasmo_4-0|5.0]]</sup> <sup>[[#cite_ref-rasmo_4-1|5.1]]</sup> Chamberlain P, Delker S, Pagarigan B, Mahmoudi A, Jackson P, Abbasian M, Muir J, Raheja N, Cathers B. Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes. PLoS One. 2014 Aug 11;9(8):e103638. doi: 10.1371/journal.pone.0103638., eCollection 2014. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/25111382 25111382] doi:[http://dx.doi.org/10.1371/journal.pone.0103638 http://dx.doi.org/10.1371/journal.pone.0103638]</li> <li id="cite_note-rasmo6-5">↑ <sup>[[#cite_ref-rasmo6_5-0|6.0]]</sup> <sup>[[#cite_ref-rasmo6_5-1|6.1]]</sup> Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A. 2007 Jan 23;104(4):1272-7. Epub 2007 Jan 16. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17227859 17227859] doi:[http://dx.doi.org/10.1073/pnas.0610251104 http://dx.doi.org/10.1073/pnas.0610251104]</li></ol></ref>