Student Project 5 for UMass Chemistry 423 Spring 2015
From Proteopedia
Contents |
LRRK2/Kinase Inhibitors
by Nick Barberio, Nicole Garvin, Megan Greiner, Peter Kelly, Charit Tippareddy, John Vetrano
Student Projects for UMass Chemistry 423 Spring 2015
Introduction
|
Parkinson’s disease is one of the most common neurodegenerative diseases worldwide. In comparison, Parkinson’s is ranked just below Alzheimer’s disease, with a 2% lifetime risk. This disorder causes neurons in the body to change their structure dramatically and lose function. These changes trigger symptoms that affect a person’s motor skills, which can include tremors, rigid muscles, slowed movement, change of speech, and weakened balance. While Parkinson’s disease is known for its prevalence with motor skills, it can also affect judgment, decision-making, and memory. Currently there are treatments to manage this disease, but a lot of progress needs to be made to find a cure [1].
Leucine rich repeat kinase 2 (LRRK2) is a key protein that can be useful in the treatment of Parkinson’s disease, but currently researchers do not fully comprehend how it specifically correlates to treatment. However, the majority of cases have a similar mutation, G2019S , in the kinase domain of LRRK2, which has proven to increase kinase activity. The idea is to detect kinase inhibitors that can reduce the activity of this mutation in hopes that it will lead towards a better treatment. A kinase is an enzyme that activates phosphate groups to transition to a substrate from high-energy molecules. In order to find more information about the kinase crystal structure of LRRK2, tyrosine kinase 2 () is used as a model because it is 74% similar in ATP-binding residues to LRRK2 [2].
Although, the kinase domain does seem the most relevant to the treatment of Parkinson’s disease, the ROC domain of LRRK2 has proven to regulate kinase activity as well. It does this by yielding GDP from GTP through alternations in its conformation in a GTP-bound and GDP-bound cycle. The GDP-Mg2+ ligand in the ROC domain plays an important role in the complex by stabilizing GDP [3].
Amino to Carboxy Rainbow (N->C Rainbow) color key for protein chains in TYK2 (pdb code 4py1)
- current model for kinase domain of LRRK2
Amino Terminus Carboxy Terminus
Overall Structure
|
The Overall Structure of TYK2 is shown in the window on the right and may be returned to by clicking .
- Secondary Structure
The protein consists primarily of alpha helices. There are 9 complete helices and 5 small helical-like structures in the protein. to view the alpha helices only. Most helices are clustered in one area, with the exception of 1 full helix and a partial helical loop.
Additionally, there are 11 beta sheets that are distributed in 3 groups among the protein. to see only the beta sheets . There is a group of 5 relatively large beta sheets adjacent to 2 medium sized beta sheets. These 2 groups make up most of the “beta region” of the protein. There are another 4 small beta sheets on the opposite end of the protein in the “alpha region”.
to combine the alpha helices and the beta sheets and begin building the protein.
Now to add the connecting loops.
Finally, to add the Ligand
- Polar and Non-polar Groups
to view the entire protein with polar and non-polar coloring.
When looking at the , you see that most residues are polar and there are fewer non-polar .
Conversely, the consist almost entirely of non-polar groups. Because the non-polar groups are buried, the protein minimizes the amount of interactions between water and hydrophobic groups. Water molecules are free to hydrogen bond and move freely around the surface of the protein. If non-polar groups were not buried, then water would form an unfavorable low entropy cage structure around the protein. Like many proteins, burying hydrophobic groups creates a stable overall structure.
- Location of Hinge Structure
There is one critical strand that connects the "alpha region" to the "beta region". This may act as a hinge to change the angle between the two regions and increase or decrease exposure to the ligand during binding interactions.
Binding Interactions
|
As a kinase, LRRK2 catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules (ATP, GTP) to specific substrates. In order to do so, LRRK2 binds to and stabilizes the transition state of ATP or GTP to allow the dephosphorylation of these molecules. Kinase recognition of correct substrates is complex and still poorly understood, however specific binding interactions between LRRK2 and several successful kinase inhibitors have been documented.
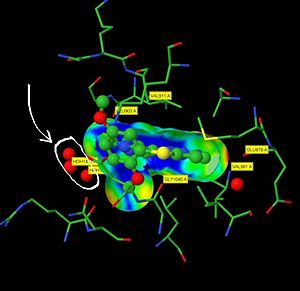
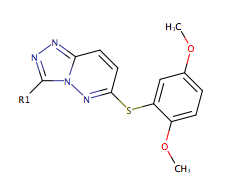
The to the right highlights the inhibition of the kinase domain of LRRK2 by 6-((2,5-dimethoxyphenyl)sulfanyl)-3-(1-methyl-1H-pyrazol-4-yl)(1,2,4)triazolo(4,3-b)pyridazine which we will refer to by its PDB code, 2YK. This molecule was shown to inhibit the kinase domain of LRRK2 by altering the conformation of the protein. This inhibitor binds to an active site pocket in the middle of LRRK2 [2]. The residues involved in binding interactions are: LEU 903, VAL 911, GLY 1040, GLU 979, VAL 981 [2].
The binding pose of this molecule within LRRK2 leaves a small section of the ligand exposed to solvent. This exposed area available to solvent can be seen in yellow in the green scene to the left. This allows for hydrogen bonding stabilization of this subsection of the ligand that is not directly interacting with residues inside LRRK2[2].
The image to the left summarizes the binding interactions between LRRK2 and 2YK. The hydrogen bonds with solvent at the exposed surface are represented as red spheres. These are circled and indicated with an arrow.
14-3-3 Interactions
14-3-3 proteins are a family of conserved regulatory molecules that are expressed in all eukaryotic cells [4]. 14-3-3 proteins have the ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors[4]. Phosphorylation of LRRK2 at Ser910 and Ser935 mediates interaction with 14-3-3[5]. These residues are highlighted in green in the scene.
Additional Features
|
LRRK2 is a very large protein comprised of multiple domains. The most well-known LRRK2 mutation occurs in the kinase domain and plays a role in Parkinson’s disease by increasing LRRK2's kinase activity.
Researchers have since discovered that the also regulates LRRK2’s kinase activity by functioning as a GTPase which switches between an active GTP- and inactive GDP-bound conformation[6]. The for both monomers is a Mg2+ binding pocket located on the surface of the ROC dimer, each shown with a bound GDP. Although the mechanism is unclear, LRRK2 is believed to exhibit intramolecular regulation: the activity of LRRK2’s kinase domain is stimulated by the active GTP-bound state of its own ROC domain.
whose mutations are associated with Parkinson’s disease, R1441 and I1371, are the key players of LRRK2’s GTPase-regulated kinase activity, because they work together to stabilize the ROC dimer [3]. Specifically, the guanidine group of R1441 exhibits essential with the backbone carbonyl oxygen of residue F1401 and the hydroxyl group on the other monomer’s nearby alpha helix. Additionally, the guanidine group, W1434’s six membered-ring, and the side chain rings of F1401 and P1406 create a via stacking interactions. The I1371 residues are located within a and provide optimal with the T1404 methyl group.
Due to the intricate hydrogen bonding, hydrophobic contacts, and other important interactions contributed by R1441 and I1371, mutations of either residue can easily destabilize the ROC dimer and alter its function. Research demonstrates that these mutations disrupt the hydrolysis of GTP to GDP, prolonging the active GTP-bound state[7]. Ultimately, this reduction in GTPase function of the mutated ROC dimer increases LRRK2’s kinase activity, a classic marker of Parkinson’s disease.
Quiz Question 1
|
The molecule shown is TYK2. Since the structure of TYK2 is almost identical to the structure of the kinase region in LRRK2, we will use it to show the concept of the .
The is the only segment that connects the (mainly) to the (mainly) . Various intramolecular interactions between the upper and lower segments help the protein maintain its tertiary structure. The strength of the interactions determines the rigidity of the hinge. If we were to add several cysteine molecules that were able to form disulfide bonds between the upper and lower segments without disrupting the active site, how would the kinase be affected? In your description, include whether the mutations would primarily affect Km or Vmax, how they would affect these kinetic parameters, and give a brief explanation of why.
Quiz Question 2
|
To the right is a green scene displaying the of LRRK2 with the residues involved in binding highlighted along with the ligand. The oxygen atoms are shown in red and the nitrogen atoms are shown in blue. Below is the molecular structure of the ligand taken from Galatsis[2] with one of the ends substituted for R. By looking at the molecular structure presented and considering the binding pocket propose three modifications at the R group which you think would increase the ligands ability to inhibit the kinase.
See Also
- [2ZEJ]
- [Leucine-rich Repeat]
- [Crystal structure of a leucine-rich repeat protein]
- [Leucine-rich repeat-contain protein 4]
- [[1]]
Credits
Introduction - Megan Greiner
Overall Structure - Nick Barberio
Drug Binding Site - John Vetrano
Additional Features - Nicole Garvin
Quiz Question 1 - Charit Tippareddy
Quiz Question 2 - Peter Kelly
References
- ↑ Schapira AH. Neurobiology and treatment of Parkinson's disease. Trends Pharmacol Sci. 2009 Jan;30(1):41-7. doi: 10.1016/j.tips.2008.10.005. Epub , 2008 Nov 29. PMID:19042040 doi:http://dx.doi.org/10.1016/j.tips.2008.10.005
- ↑ 2.0 2.1 2.2 2.3 2.4 Galatsis P, Henderson JL, Kormos BL, Han S, Kurumbail RG, Wager TT, Verhoest PR, Noell GS, Chen Y, Needle E, Berger Z, Steyn SJ, Houle C, Hirst WD. Kinase domain inhibition of leucine rich repeat kinase 2 (LRRK2) using a [1,2,4]triazolo[4,3-b]pyridazine scaffold. Bioorg Med Chem Lett. 2014 Sep 1;24(17):4132-40. doi: 10.1016/j.bmcl.2014.07.052., Epub 2014 Jul 30. PMID:25113930 doi:http://dx.doi.org/10.1016/j.bmcl.2014.07.052
- ↑ 3.0 3.1 Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci U S A. 2008 Feb 5;105(5):1499-504. Epub 2008 Jan 29. PMID:18230735
- ↑ 4.0 4.1 Bartel M, Schafer A, Stevers LM, Ottmann C. Small molecules, peptides and natural products: getting a grip on 14-3-3 protein-protein modulation. Future Med Chem. 2014 May;6(8):903-21. doi: 10.4155/fmc.14.47. PMID:24962282 doi:http://dx.doi.org/10.4155/fmc.14.47
- ↑ Mamais A, Chia R, Beilina A, Hauser DN, Hall C, Lewis PA, Cookson MR, Bandopadhyay R. Arsenite stress down-regulates phosphorylation and 14-3-3 binding of leucine-rich repeat kinase 2 (LRRK2), promoting self-association and cellular redistribution. J Biol Chem. 2014 Aug 1;289(31):21386-400. doi: 10.1074/jbc.M113.528463. Epub, 2014 Jun 18. PMID:24942733 doi:http://dx.doi.org/10.1074/jbc.M113.528463
- ↑ Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007 Oct 1;313(16):3658-70. Epub 2007 Jul 19. PMID:17706965 doi:http://dx.doi.org/10.1016/j.yexcr.2007.07.007
- ↑ Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem. 2007 Oct;103(1):238-47. Epub 2007 Jul 10. PMID:17623048 doi:http://dx.doi.org/10.1111/j.1471-4159.2007.04743.x