Student Project 6 for UMass Chemistry 423 Spring 2015
From Proteopedia
Contents |
Human Pim-1 Kinase in Complex with an Aminooxadiazole-indole Inhibitor - 4ty1
by Ashley Andrews, Steven Ayotte, Lee Johnson, Deanna Oliveira, Seo Yeon Orite, Mark Allen Pagkaliwangan
Student Projects for UMass Chemistry 423 Spring 2015
Introduction
|
In many forms of cancer, the disease arises from a mutation in the cell that blocks the regulation pathways of cell replication. First discovered in the murine leukemia virus, the proviral integration sites of Moloney (Pim) follows a kinase pathway that is built mostly upon nonspecific serine and threonine kinases [1].
The Pim-1 kinase is critical in cell proliferation and signaling pathways as the cell is undergoing replication. When a cell is damaged and labeled unfit to continue in the replication process, it goes through a process of apoptosis, or cell death. Pim-1 is able to bypass apoptosis by phosphorylating the apoptosis signaling kinase 1 (ASK1) which induces apoptosis, therefore increasing the likelihood of cell survival.
This process is evolutionarily beneficial for when cells are in a high stressed environments and would not be able to survive under strictly regulated replication parameters. However, extreme over expression of Pim-1 allows for multitudes of damaged cells to thrive, which is the fundamental basis for cancer development. Pim-1 over expression is seen in many cancerous diseases including multiple myeloma,4 acute myeloid leukemia, prostate cancer, a gastric and liver carcinomas, and autoimmune diseases. In studies, it is theorized that inhibiting the Pim-1 kinase could have value as a therapeutic drug to reduce damaged cell replication.
Pim-1 has two ligands, and which are the primary source of binding with ASK1. One useful function of 38W is that it has a proline residue in the hinge, permitting it to create only one hydrogen bond with ATP. This allows for selectivity for the Pim-1 kinase .
Overall Structure
|
Secondary Structure
This kinase is a transferase inhibitor with a 1-chain structure. This single chain contains both alpha helices and beta strands. There are 15 alpha helices and 12 beta strands. The alpha helices are clumped together on one side of the molecule and the beta strands are together on the other side. If the strand were laid out linearly, the alpha helices would be linked together on one side of the chain and the beta strands would be linked together on the other.[2] In the to the right, the alpha helices are blue and the beta strands are green. This protein also contains two ligands, 38W and GOL. The ligand 38W, which is black in the figure, is somewhat hidden inside of the molecule, showing that it contains interactions within the molecule. The ligand GOL is placed on the edge of the protein, allowing for outside interactions along with interactions inside the molecule.
Acidity and Polarity
This molecule has a dipole moment between the acidic and basic, or negatively and positively charged portions of the molecule. , the acidic/negative areas are purple, the basic/positive areas are green, and the neutral areas are golden brown. These basic and acidic areas of the molecule alternate, just like the polar and non polar segments of chain alternate.
, the polar regions are purple and the nonpolar regions are pink. The hydrophobic areas of the molecule correlate with the pink non-polar regions in this figure. These regions alternate throughout the entire chain and show that there is no relationship between polarity and secondary structure here.
Binding Interactions
|
4ty1 contains two ligands, glycerol(GOL) and N-tert-butyl-5-[3-(4-cyclopropylpyrimidin-2-yl)-1H-indol-5-yl]-1,3,4-oxadiazol-2-amine(38W). 38W is the aminooxadiazole-indole inhibitor compound which inhibits the PIM-1 kinase. It inhibits this kinase by competatively binding to its ATP binding site. It binds to the protein using the following interctions.
38W Aminooxadiazole Ligand Binding Interactions

-Hydrogen Bonding Interactions:
- 3.08 Å H-bond between 38W indole N-H and Glu 121 carbonyl group.
- H-bonding between 38W oxadiazole ring N-H and Glu89 carboxyl (3.0 Å) & 38W oxadiazole ring N and Lys67 N-H (2.92 Å).
- 3.4 Å H-bonding between 38W tert-butylamino group and Asp186.
-Van der Waals Force Interactions:
- potential van der waals interactions between 38W benzene ring and Leu 120.
-Hydrophobic Contacts:
- hydrophobic contact between 38W pyrimidine ring and Val126, Leu174, and Ile185.
*based on research by Wurz et al.
[3]
Additional Features: Inhibition of ATP and Pim-1 Kinase Binding
|
Pim-1 is constitutively active. Being always active, its activity level solely depends on the absolute amount of protein present in the cell [4]. Therefore, Pim-1 activity is normally regulated at the transcription or translation level. In addition, Pim-1 needs to be removed regularly for cells to function normally. For a cancer therapy, however, Pim-1 is being regulated via post-translational method by using small molecule inhibitor that prevents Pim-1 from phosphorylating other proteins involved in cancer development. Post-translational method is preferred over gene therapy because inhibitors are less invasive treatment that only requires oral intake.
All reported small molecule Pim-1 inhibitors are known to competitive with ATP to bind with Pim-1 kinase. This competitive binding can occur via two different methods. One method is by closely mimicking the ATP binding to the hinge region of the kinase. Another method is using other areas of the active site to bind with the kinase [5]. The latter method still prevents ATP from binding with Pim-1.
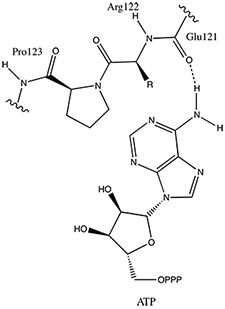
As shown in figure, ATP binds with the hinge region of the kinase. This hinge region lacks hydrogen bond donor and, therfore, can easily form hydrogen bond with ATP. [6]. Primary amine group of ATP forms a hydrogen bond with the carbonyl oxygen of Glu 121. Knowing this simple interaction between ATP and Pim-1 kinase, scientists studied many small molecules that target this interaction. Aminooxadiazole-indole, the inhibitor of interest, was also designed to target this specific interaction as shown in this . Aminooxadiazole not only forms a stable hydrogen bond with Glu 121, but it also interacts with other side chains around of Pim-1. Binding to the kinase at the hinge region and other active sites, aminooxadiazole-indole inhibitor outcompetes ATP and effectively inactivates Pim-1.
Quiz Question 1
|
This highlights the GLU 121A and LYS 67A residues in the 38W binding site (differentiated by their ball and stick representations, where red=O, blue=N). By what mechanism do these residues stabilize the ligand? Click and scroll to zoom in on the figure.
Quiz Question 2
|
In its active state, ATP binds with Glu 121 in the hinge region of the kinase. Assuming PIM-1 follows Michaelis–Menten kinetics, if we study the reaction rates of PIM-1 with and without 38W, what do you expect to find? How will Vmax change? How about Km? To maximize inhibitory efficiency, do we want a large change in Km or a small change in Km?
See Also
Credits
Introduction - Deanna Oliveira
Overall Structure - Ashley Andrews
Drug Binding Site - Steven Ayotte
Additional Features - Seo Yeon Orite
Quiz Question 1 - Mark Allen Pagkaliwangan
Quiz Question 2 - Lee Johnson
References
- ↑ Gu JJ, Wang Z, Reeves R, Magnuson NS. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene. 2009 Dec 3;28(48):4261-71. doi: 10.1038/onc.2009.276. Epub 2009 Sep 14. PMID:19749799 doi:10.1038/onc.2009.276
- ↑ "Serine/threonine-protein Kinase Pim-1." PIM1_HUMAN. UniProt Consortium, 2015. Web. 03 Apr. 2015. <http://www.uniprot.org/uniprot/P11309>.
- ↑ Wurz RP, Pettus LH, Jackson C, Wu B, Wang HL, Herberich B, Cee V, Lanman BA, Reed AB, Chavez F Jr, Nixey T, Laszlo J 3rd, Wang P, Nguyen Y, Sastri C, Guerrero N, Winston J, Lipford JR, Lee MR, Andrews KL, Mohr C, Xu Y, Zhou Y, Reid DL, Tasker AS. The discovery and optimization of aminooxadiazoles as potent Pim kinase inhibitors. Bioorg Med Chem Lett. 2015 Feb 15;25(4):847-55. doi: 10.1016/j.bmcl.2014.12.067. , Epub 2015 Jan 7. PMID:25599837 doi:http://dx.doi.org/10.1016/j.bmcl.2014.12.067
- ↑ Wurz, R. P.; Pettus, L. H.; Jackson, C.; Wu, B.; Wang, H.; Herberich, B.; Cee, V.; Lanman, B. A.; Reed, A. B.; Chavez Jr., F.; Nixey, T.; Laszlo III, J.; Wang, P.; Nguyen, Y.; Sastri, C.; Guerrero, N.; Winston, J.; Lipford, J. R.; Lee, M. R.; Andrews, K. L.; Mohr, C.; Xu, Y.; Zhou, Y.; Reid, D. L.; Tasker, A. S. The discovery and optimization of aminooxadiazoles as potent Pim kinase inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 847-855.
- ↑ Merkel, A. L.; Meggers, E.; Ocker, M. PIM1 kinase as a target for cancer therapy. Expert Opin. Investig. Drugs 2012, 21, 425-436.
- ↑ Alex N. Bullock, Judit Debreczeni, Ann L. Amos, Stefan Knapp, and Benjamin E. Turk Structure and Substrate Specificity of the Pim-1 Kinase J. Biol. Chem. 2005 280: 41675-41682. First Published on October 13, 2005, doi:10.1074/jbc.M510711200
