Student Project 7 for UMass Chemistry 423 Spring 2015
From Proteopedia
Contents |
MTH1/crizotinib
by Colin Hannahan, Megi Marina, Cassandra Martin, Eric Rice, Benjamin Ryter, Matthew Tuttle
Student Projects for UMass Chemistry 423 Spring 2015
Introduction
|
MTH1 is known as 7,8-dihydro-8-oxoguanine triphosphatase (2). MTH1 is a protein in the human body encoded by the NUDT1 gene. In general, MTH1 is often synonymous for the gene NUDT1.The protein belongs to the Nudix hydrolase family, and is characterized by a conserved of 23 – residue sequence segment. MTH1 is part of the Nudix hydrolyse family, it has the typical Nudix structure, fold, structure arranged as a mixed of with one on each side.
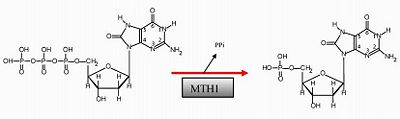
MTH1 prevents the misincorporation of oxidized nucleotides into DNA. Reactive oxygen species (ROS) produced as byproducts from cellular metabolism can cause damage to DNA and free nucleotides. Misincorporation of oxidized nucleoside triphosphates into DNA and RNA is one cause of mutation during transcription and replication that can result in DNA damage and ultimately cell death. MTH1 performs the hydrolysis of a triphosphate nucleotide to a monophosphate nucleotide to prevent this incorporation. The hydrolysis of the triphosphate nucleotide 8-oxo-dGTP to the monophosphate nucleotide 8-oxo-dGMP will not be recognized by DNA polymerase (1). This misincorporation of oxidized nucleotides could be used as a tool to combat tumor growth. Cancer cells frequently overexpress MTH1, however; suppressing this protein could cause an increase in DNA misincorporations and induce cell death in tumors.
MTH1 is inhibited by . Crizotinib has an aminopyridine structure and functions as a protein kinase inhibitor because it binds with the ATP – binding pocket of target kinase (3). Crizotinib has a high impact in drug design because it causes tumors to shrink and stabilize. It has been shown that when (S)-Crizotinib inhibits MTH1, DNA single–strand breaks are more prevalent and DNA repair in colon carcinoma cells is activated. When (S)-Crizotinib inhibits MTH1, the protein can no longer function as a nucleotide ‘healer’, and the impairment rate of the nucleotide pool is increased. One study showed MTH1 impairs growth of KRAS tumor cells and an overexpression of MTH1 mitigates the cancer cells(4). Targeting MTH1 and other enzymes which control the cleansing of oxidized nucleotides as a tumor-specific strategy may represent a novel strategy for difficult-to-treat tumors.
Overall Structure
|
MTH1/S-crizotinib is a member of a protein family of phosphorohydrolases known as the Nudix family (2). Proteins in the Nudix family are used to hydrolyze nucleoside phosphates; they also act as catalysts for the same reaction. Proteins in the Nudix family consist of two unique parts: (2). The Nudix fold is the secondary structure, or backbone, of the protein; the backbone has an α/β/α structure, meaning that the backbone consists of mixed beta sheet (gold) with an alpha helix on each end (magenta) (2).
The second component of proteins in the Nudix family, such as MTH1/S-crizotinib, is the Nudix motif, which is different for every protein. The Nudix motif in MTH1/S-crizotinib is made up of 23 amino acid sequence that orient into a small alpha helix (dark purple) (5,6). This helix attached to one of the main alpha helices in MTH1/S-crizotinib’s backbone. The amino acids that catalyze the hydrolysis reactions are located in this alpha helix (2,5,6). All of these components establish the of the protein; the outer parts of the helices and beta sheet are polar and hydrophilic (purple), while the inner parts and non polar and hydrophobic (gray).
MTH1/S-crizotinib also has an that is formed between the beta sheet and one of the alpha helices (2). This pocket is built out of the residues Leu9 (aqua), Phe27 (violet), Phe72 (magenta), Met81 (lime green), Val83 (brown), Trp117 (orange), Trp123 (gold), and Phe139 (salmon). Furthermore, the amino acid (magenta) is used to coordinate metal-binding in the protein and is located outside the alpha helix motif (2).
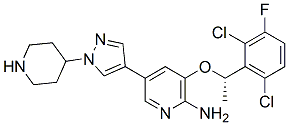
MTH1/S-crizotinib has three different types of ligands attached to it: one chloride ligand, five sulfate ligands, and the . Crizotinib is a tyrosine kinase inhibitor, and its molecular structure is C21H22Cl2FN5O. The stereochemistry of the crizotinib determines if this inhibitor prevents MTH1 from carrying out its purpose in cancer cells; the S-crizotinib stops MTH1 from working while the R-crizotinib has no effect on the protein (3,7).
Binding Interactions
|
Human MutT homologue, also known as MTH1, is a nucleotide pool sanitization enzyme and the target for crizotinib, a competitive inhibitor. Crizotinib is a kinase inhibitor that suppresses MTH1 activity in concentrations as low as nanomoles/liter, suggesting high levels of specificity (1). Crizotinib acts on MTH1 through competitive inhibition, meaning it displaces kinases, the usual substrates. Because the specificity of the binding pocket of MTH1 depends on the stabilization of the enol tautomer of 8-oxo-dGTP, crizotinib’s chemistry interacts with this conformation to be effective (2). The binding of crizotinib yields no major structural changes in the MTH1 protein.
The most significant interactions between crizotinib and the MTH1 active site are hydrogen bonding interactions. Hydrogen bonding occurs between the Asparagine residue 33 and the N3 of 8-oxo-dGMP and between the acidic Aspartate residues 119 and 120 and the oxygen in 8-oxo-dGMP. Both the Asp 119 residue and the Asn 33 residue are crucial to binding specificity (2). Because crizotinib has a chiral center, the (S) and (R) enantiomers act differently in the binding pocket of MTH1. After recent testing of MTH1 using both enantiomers, the (S) enantiomer has shown a higher affinity for the MTH1 binding pocket, whereas the (R) enantiomer is a better inhibitor for most other similar protein kinases. An eclipsed conformation of a methyl group at the chiral center and a chlorine attached to the benzyl ring are responsible for reducing the energetic favorability of the (R) enantiomer in the binding pocket of MTH1 (1).
Additional Features
|
The can be seen inside the globular protein structure. The majority of the residues appear exposed on the protein surface rather than buried within the protein. Acidic and basic residues are also prevalent within the binding pocket.
MTH1 structurally recognizes and distinguishes 8-oxo-dGTP from 8-oxo-dGMP different from one another. In order to understand how MTH1 functions, this is essential to explore. The protein MTH1 has been co-crystallized with the substrate 8-oxo-dGTP and the structure of the crystal was observed (2). A reaction took place and the product, 8-oxo-dGMP, was bound in a pocket formed by the . This green scene representation is not exact, and a structural change may have occurred upon 8-oxo-dGTP binding. make hydrogen bonds to the 2-amino group of 8-oxo-dGMP. This amino position is central for the interaction. Nucleotide analogs were used to test MTH1 substrate recognition; results were in agreement. Removal of the 2-amino group led to a 10-fold less effective degradation (2). Mg2+ also appeared in the co-crystal structure, but the electron density, coordination geometry, and coordination distances showed Mg2+ was not bound to the complex. It was hypothesized that Mg2+ may coordinate with the substrate of the triphosphate nucleotide (2). Many other residues involved in specificity and recognition have been identified.
MutT is a homolog of MTH1, shares the same nubix family, and is expressed in Escherichia coli. MutT also catalyzes the hydrolysis of 8-oxo-dGTP to 8-oxo-dGMP, but has a 21% (low) sequence identity to MTH1. Co-crystal structure produced the anti conformation of bound 8-oxo-dGMP in MTH1 while syn conformation was observed in MutT (2). From the crystal structure it was determined that the base–protein interactions are quite different and reveal no conservation of structure. Although the proteins shared a conserved catalytic region, the nucleotide recognition was not conserved. Diverging evolutionary routes have been taken, and one possibility is to allow MTH1 to evolve more non-specific substrate specificity (2). Understanding the mechanisms of non-specific substrate binding could be an important step to developing new inhibitors for similar enzymes.
Quiz Question 1
|
- This scene colors the protein from the N-terminus to the C terminus.
- This scene shows the active site of the protein. Individual amino acids are labelled and active site residues are colored according to charge (red = positive, blue = negative, white = uncharged). The ligand (S-crizotinib) in the center is colored by atom (Black = Carbon, Red = Oxygen, Blue = Nitrogen, Green = Halogen).
Using the green scenes provided above, answer the following:
a.) Suppose there is a point mutation causing a change at ASP-120 in MTH1. Which of the following amino acids substitutions would be least likely to affect substrate binding? Explain your reasoning.
1.) Lysine (LYS)
2.) Glutamate (GLU)
3.) Glycine (GLY)
b.) If there were a point mutation at PHE-27, which of the following amino acid substitutions would likely affect ligand binding the most? Again, explain your reasoning.
1.) Tryptophan (TRP)
2.) Tyrosine (TYR)
3.) Arginine (ARG)
Quiz Question 2
The above two green scenes highlight the chiral center of the MTH1 inhibitor called crizotinib. Research has shown that the (S)-enantiomer binds with more affinity to it's MTH1 substrate but that it is not due to interaction with the protein itself at the binding site.
|
|
Using the green scenes and the structure image can you see an unfavorable interaction near the chiral center in the (R)-enantiomer that does not exist in the (S)-enantiomer? Explain.
The conformation of the enantiomer is ___________ unfavored.
See Also
- MTH1 (7,8-dihydro-8-oxoguanine triphosphatase)
Credits
- Introduction - Megi Marina
- Overall Structure - Cassandra Martin
- Drug Binding Site - Ben Ryter
- Additional Features - Eric Rice
- Quiz Question 1 - Matt Tuttle
- Quiz Question 2 - Colin Hannahan
References
- Huber, Kilian V. M., Eidarus Salah, Branka Radic, Manuela Gridling, Jonathan M. Elkins, Alexey Stukalov, Ann-Sofie Jemth, Camilla Göktürk, Kumar Sanjiv, Kia Strömberg, Therese Pham, Ulrika Warpman Berglund, Jacques Colinge, Keiryn L. Bennett, Joanna I. Loizou, Thomas Helleday, Stefan Knapp, and Giulio Superti-Furga. "Stereospecific Targeting of MTH1 by (S)-crizotinib as an Anticancer Strategy." Nature 508 (2014): 222-27. Web.
- Svensson, Linda M., Ann-Sofie Jemth, Mathhieu Desroses, Olga Loseva, Thomas Helleday, Martin Higbom, and Pal Stenmark. "Crystal Structure of Human MTH1 and the 8-oxo-dGMP Product Complex." Http://www.sciencedirect.com. N.p., 19 Aug. 2011. Web.
- "Crizotinib | C21H22Cl2FN5O - PubChem." Crizotinib | C21H22Cl2FN5O - PubChem. N.p., n.d. Web. 03 Apr. 2015.
- Fricker, Janet. "Two Studies Show MTH1 Offers Promising New Target for Cancer Treatment." . Ecancer. Cancer Intelligence, 02 Apr. 2014. Web. 03 Apr. 2015.
- M. Mishima,; Y. Sakai,; N. Itoh,; H. Kamiya,; M. Furuichi,; M. Takahashi, Y. Yamagata, S. Iwai,; Y. Nakabeppu,; and M. Shirakawa. “Structure of the Human MTH1, a Nudix Family Hydrolase that Selectively Degrades Oxidized Purine Nucleoside Triphosphates”, J Biol Chem., 279, no. 32, 33806-33815, 2004.
- S.B. Gabelli,; M.A. Bianchet,; M.J. Bessman,; L.M. Amzel “The structure of ADP-ribose pyrophosphates reveals the structural basis for the versatility of the Nudix family”, Nat Struct Biol, 8, 467-472, 2001.
- "Crizotinib, (S)-." (S)-Crizotinib. N.p., n.d. Web. 03 Apr. 2015.