Function
Transcription activator-like effector PthXo1 (TALE PthXo1) TALEs are proteins originally derived from the plant pathogene Xanthomonas spp. The gene encoding for the effector are Pthxo1 and Pxo_00227. Originally they were detected in Xanthomonas Oryzae but the TALE PthXo1 genes were transformed into Escherichia Coli to express the DNA-binding protein.
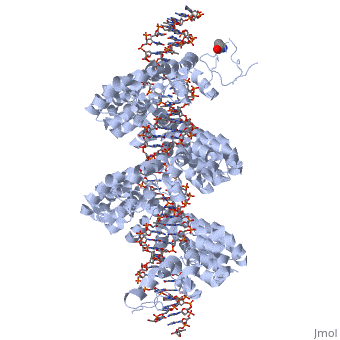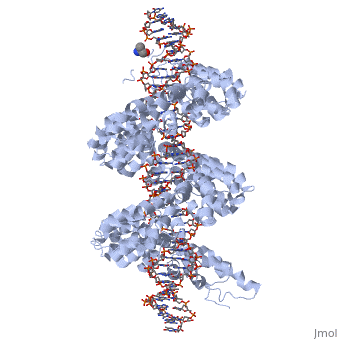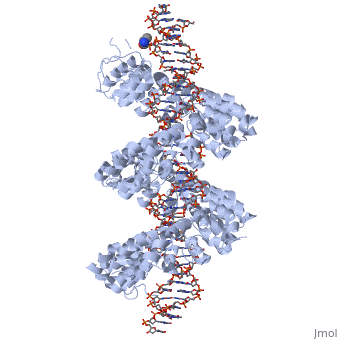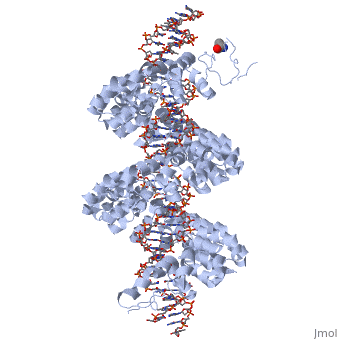
DNA recognition by TAL effectors is mediated by tandem repeats, each 33 to 35 residues in length, that specify nucleotides via unique repeat-variable diresidues (RVDs). The crystal structure of PthXo1 bound to its DNA target was determined by high-throughput computational structure prediction and validated by heavy-atom derivatization. Each repeat forms a left-handed, two-helix bundle that presents an RVD-containing loop to the DNA. The repeats self-associate to form a right-handed superhelix wrapped around the DNA major groove. The first RVD residue forms a stabilizing contact with the protein backbone, while the second makes a base-specific contact to the DNA sense strand. Two degenerate amino-terminal repeats also interact with the DNA. Containing several RVDs and noncanonical associations, the structure illustrates the basis of TAL effector–DNA recognition [1]
The interaction between the and a is displayed on the right side.
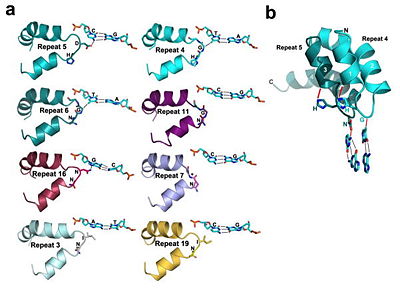
Fig.1 Structure of the PthXo1 DNA binding region in complex with its target site. The coloring of individual repeats matches the schematic in XXX
DNA binding module with marked RVD
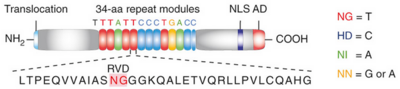
Figure 2: Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = , HD = , NI = , and NN = or . The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site.[2]
Tertiary structure of the RVD
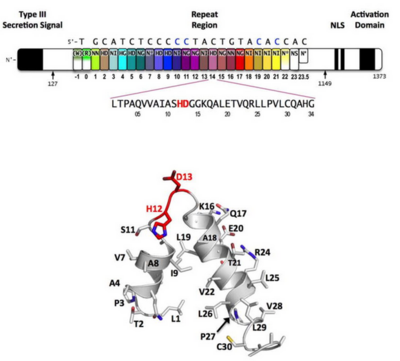
Figure 3: Domain organization of PthXo1 and structure of a single TAL effector repeat
TAL effectors contain N-terminal signals for bacterial type III secretion, tandem repeats that specify the target nucleotide sequence, nuclear localization signals, and a C-terminal region that is required for transcriptional activation. PthXo1 contains 23.5 canonical repeats (color coded to match Figure 3) that contact the DNA target found in the promoter of the rice Os8N3 gene (17). Blue bases correspond to positions in the target where the match between protein and DNA differs from the optimal match specified by the recognition code (3,4). Arrows indicate the start and end of the crystallized protein construct. In the structure, repeats 22 to 23.5 are poorly ordered, as are the C-termini of the two N-terminal cryptic repeats. The sequence and structure of a representative repeat (#14) is shown; RVD residues (HD) that recognize cytosine are red.[3]
Figure 4: All of the repeats in the DNA-bound PthXo1 structure form highly similar two-helix bundles (Figure 1c). The helices span positions 3 to 11 and 14 to 33, locating the RVD in a loop between them. A proline located at position 27, creates a kink in the second helix that appears to be critical for the sequential packing and association of tandem repeats with the DNA double helix. The packing of consecutive helices within and between individual repeats is left-handed, in contrast to the right-handed packing of helices found in TPR proteins (10). The modular architecture of the TAL effector repeats is reminiscent of the mitochondrial transcription terminator mTERF (11) and the RNA-binding attenuation protein TRAP (12).
Sequence-specific contacts of PthXo1 to the DNA are made exclusively by the second residue in each RVD to the corresponding base on the sense strand. In contrast, the side chain at the first position of each RVD contacts the backbone carbonyl oxygen of position 8 in each repeat, constraining the RVD-containing loop (Figure 3). Additional, nonspecific contacts to the DNA are made by a lysine and glutamine found at positions 16 and 17.
Interaction between RVDs and DNA
‘HD’ RVDs:
the aspartate residue makes van der Waals contacts with the edge of the corresponding cytosine base and a hydrogen bond to the cytosine N4 atom.
‘NG’ and ‘HG’ RVDs:
make a contact in which the backbone alpha carbon of the glycine residue forms a nonpolar van der Waals interaction with the methyl group of the opposing thymine base (average distance ~ 3.3 Å). At the one position where an NG is aligned opposite a cytosine base, the backbone carbonyl and alpha-carbon of the same glycine residue displays a less favorable, far more distant contact (~ 6 Å).
‘NN’ RVDs:
is positioned to make a hydrogen bond with the N7 nitrogen of an opposing guanine base. This RVD associates with either guanosine or adenine with roughly equal frequency (3, 4, 14); the availability of an N7 nitrogen in either purine ring appears to explain that observation (13).
‘N*’RVDs:
PthXo1 contains two 33 residue (7 and 22). Since RVDs are followed immediately by two conserved glycine residues, this repeat is equivalent to an ‘NG’ repeat in which one of those glycine residues is missing. The crystal structure indicates that the deletion results in a truncated RVD loop that extends less deeply into the DNA major groove, with the glycine at position 13 located a considerable distance (over 6 Å)
‘NI’ RVDs:
Occurs seven times in PthXo1, and displays an unusual contact pattern to adenosine or cytosine bases. The aliphatic side chain of the isoleucine residue is observed to make non-polar van der Waals contacts to C8 (and N7) of the adenine purine ring, or to C5 of the cytosine pyrimidine ring.
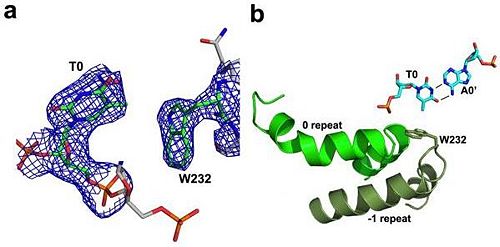
N-terminal to the canonical repeats, the PthXo1 structure reveals two degenerate repeat folds that appear to cooperate to specify the conserved thymine that precedes the RVD-specified sequence (Figure 5). We have designated these as the 0th and -1st repeats. Residues 221 to 239 and residues 256 to 273 each form a helix and an adjoining loop that resembles helix 1 and the RVD loop in the canonical repeats; the remaining residues in each region are poorly ordered. Those two N-terminal regions converge near the 5′ thymine base, with the indole ring of tryptophan 232 (in the -1st repeat) making a van der Waals contact with the methyl group of that base. Mutation of the thymine reduces TAL effector activity at the target (3, 15). Tryptophan 232, as well as the surrounding residues, is highly conserved across available, intact TAL effector sequences.