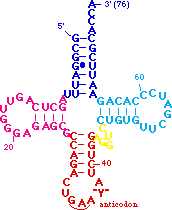Source [1]
Structural highlights
Domains
.
The acceptor stem includes the 5' and 3' ends of the tRNA. The 5' end is generated by RNaseP :-). The 3' end is the site which is charged with amino acids for translation. Some aminoacyl tRNA synthetases interact with both the acceptor 3' end and
the anticodon when charging tRNAs. from the anticodon loop, at bottom, by clicking here. Note also how the acceptor stem stacks onto the TpsiC stem to form a continuous helix. The anticodon stem also stacks onto the junction between the variable loop and the D stem to form another nearly perfect helix. The TpsiC and D loops interact to bring the "cloverleaf" secondary structure in to the L-shaped tertiary structure.
Core Tertiary Interactions
Now .
Most of the backbone is shown in ribbon format, with the same color scheme as above. Several unusual base pairs, base triples and turns are highlighted and color-coded.
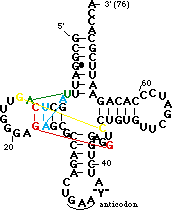
. The yellow residues are a parallel base pair (compared to the normal anti parallel) between G15 of the D-loop and C48 of the variable loop. This brings the D-loop and variable loop together. Note the sharp turn in the backbone between C48 and C49 caused by the parallel pair.
The green residues are a reverse Hoogsteen pair between U8 and A14. This pairing is important for positioning of the D stem relative to the stacked T and acceptor stems.
The cyan residues are a base triple in which A9 H-bonds in the major groove to A23 (which is paired with U12). It stabilizes a sharp turn between bases 9 and 10.
The red residues are a base triple in which 7-methyl-G46 from the variable loop H-bonds to the G22-C13 base pair of the D stem. This helps dock the variable loop onto the D-stem.
A9 is also involved in another type of tertiary interaction: it is 7-methyl-G46 and G45. In order to make room between these bases for A9 the backbone is extended by a C2' endo sugar pucker at 7-methyl-G46.
U-turns
The are responsible for turns in the anticodon and T loops. The turn is stabilized by an H-bond between a conserved U residue and the phosphate backbone and an H-bond fron the O2' of the U to the N7 of a conserved purine. . Residues 33-35 form the U-turn, N3 of U33 H-bonds to the phosphate oxygen of A36, the O2' of U33 H-bonds to the N7 of A35. Residues 34, 35 and 36 are the anticodon bases used in translation. The hypermodified wybutrosine YG37 is believed to form a steric block to frameshifting during translation.
The U-turn motif is repeated in the "T-loop" of tRNA. . Residues 55-57 form the U-turn here. A U-turn is also found in the active site of the hammerhead ribozyme.