Triosephosphate Isomerase
From Proteopedia
TRIOSEPHOSPHATE ISOMERASE (TIM, or TPI)
Triosephosphate isomerase is a key enzyme in the glycolytic pathway, which catalyzes the interconversion of dihydroxyacetone phosphate (DHAP) and (R)-glyceraldehyde-3-phosphate (GAP). The remarkable specificity and catalytic power of the enzyme has inspired intensive studies using structural biology, biophysics, and computer simulations. TIM is a catalytically perfect enzyme in the sense that its kcat/Km value is in the diffusion-limited range, and because catalytic efficiency is not improved by changes to the chemical composition of the solvent, or by changes to the amino acid sequence of the enzyme. TIM has been estimated to lower the activation energy of the reaction by 11-13 kcal/mol.
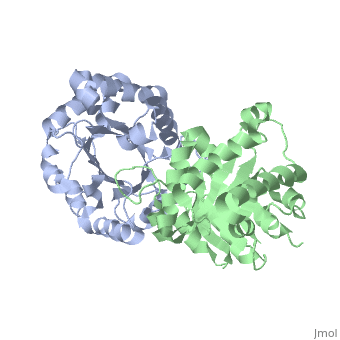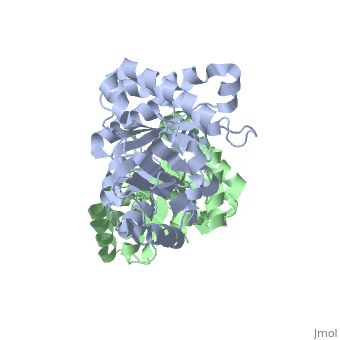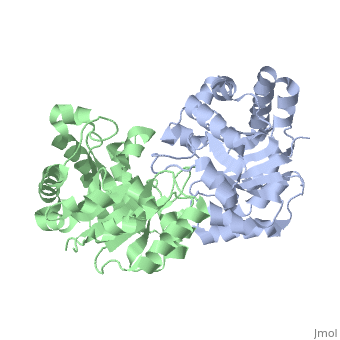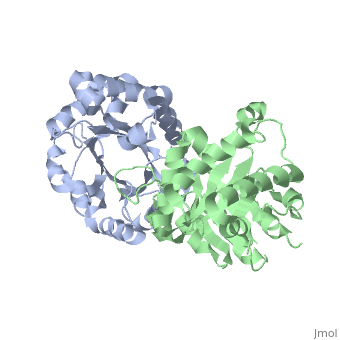
Active site features (Green links change the 3D image)The enzyme is a dimer of identical subunits. The tertiary fold of each subunit is an alpha/beta barrel of which TIM is the prototype (). Eight parallel beta strands (purple) form the wall of the barrel, which is located in the protein’s interior. Alpha helices (blue), which are connected to the beta strands, form the outer rim of the enzyme. Regions between the alpha helices and beta strands are also shown (green). The curvature of the barrel arises principally from the right-handed twist of the beta sheet. Nonpolar amino acids pointing inward from the beta strands contribute to the hydrophobic () core of the structure (grey), whereas residues pointing outward interact with the nonpolar face of the alpha helices on the outer rim (fuchsia). The is found at the carboxyl end of the barrel structure. Key active site residues include Glu165 (yellow), His95 (green), and Lys12 (blue). A flexible loop of residues 168 – 177 (highlighted in red) functions as a lid to the active site. It opens to provide access of the substrate to the active site, and it closes the active site upon substrate binding. Reaction mechanismThe reaction mechanism for the isomerization of DHAP and G3P involves acid base catalysis proceeding through an enediol intermediate [ [1] ]. The active site residues, Glu165 and His95, were shown by early crystallization studies containing bound substrate or inhibitor, to be precisely positioned to serve as catalysts of the reaction. Glu165 initiates the first step of the reaction by abstracting a proton from the pro(R) position of carbon 1 of DHAP (rate determining step) as the oxygen of carbon 2 abstracts a proton from His95. This promotes rearrangement to the enediol intermediate. In the second step, His95 abstracts a proton from the oxygen of carbon 1 as the oxygen of carbon 2 abstracts a proton from Glu165. This step promotes rearrangement to the product, G3P, thereby regenerating the enzyme. The importance of these residues in catalysis was confirmed by the kinetic analysis of site-directed mutations. For example, catalytic activity decreases by orders of magnitude when Glu165 is mutated to Asn165. Additional studies using NMR spectroscopy have demonstrated a drastically lowered pKa for His95 and formation of a hydrogen bond with the substrate analogues’ carbonyl oxygen. Lys12 also plays an important role in catalysis. This residue is highly conserved, and electrostatic interactions with the bound substrate are implicated by x-ray crystallography. As a test of its functional role, Lys12 was mutated to methionine. The Met12 mutant enzyme was found to have a defect in substrate binding despite having a correctly folded active site and overall native protein structure. Steric hindrance was not an explanation for the defect in substrate binding, whereas the electrostatic environment of the active site was significantly altered for the Met12 mutant. Thus, Lys12 contributes an appropriate electrostatic environment in the active site to favor the binding of the negatively charged substrate. Dynamic movements and role of the flexible loop in catalysisMovie 2 shows the binding of DHAP and the changes that occur during catalysis. Note the motions in Glu165 (yellow), His95 (green) and Lys12 (blue) on binding of DHAP. This movie was creating by morphing between unbound and bound TIM (PDB ID 1YPI and 2YPI, respectively), and rendered using Molscript and Raster3D. 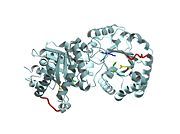 This movie shows the binding of DHAP and the changes that occur during catalysis. Also see hinge motion via Mark Gerstein's Molecular Movements database [ [2] ]. The residues in the flexible loop are highly conserved among a variety of TIM enzymes from different organisms as an indication of their functional significance. Four residues in the flexible loop were deleted by site-directed mutagenesis to test the functional role of the loop region in catalysis. The deletion mutant was found to have a severe defect in kcat despite a modest defect in Km indicating a primary role for the loop region. Furthermore, a significant increase in the level of methylglyoxal was observed for the deletion mutant compared to the wild type enzyme. Methylglyoxal is a toxic by-product of the reaction, which builds up when the enediol intermediate is released from the active site. Thus, the flexible loop also functions to hold the intermediate in the active site so catalysis can proceed to the final product. Why is the enzyme an obligate dimer?An interdigitating loop (residues 71-77) is found on each subunit. The loop extends from one subunit into a pocket near the active site of the other subunit. Notably, there is a hydrogen bond network involving Lys12 and Glu97 of one subunit and Thr75 of the other subunit. This may explain why the dimer is required for catalytic activity. Also of interestLinks to other Triosephosphate Isomerase sites [ [3] ] [ [4] ] Enzyme catalysis [ [5] ], and Enzyme kinetics [ [6] ] ReferencesBlacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988 Feb 23;27(4):1158-67. PMID: 3365378 Joseph-McCarthy D, Lolis E, Komives EA, Petsko GA. Crystal structure of the K12M/G15A triosephosphate isomerase double mutant and electrostatic analysis of the active site. Biochemistry. 1994 Mar 15;33(10):2815-23. PMID: 8130194 Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci U S A. 2005 May 10;102(19):6679-85. Epub 2005 May 3. PMID: 15870208 Komives EA, Chang LC, Lolis E, Tilton RF, Petsko GA, Knowles JR. Electrophilic catalysis in triosephosphate isomerase: the role of histidine-95. Biochemistry. 1991 Mar 26;30(12):3011-9. PMID: 2007138 Lolis E, Alber T, Davenport RC, Rose D, Hartman FC, Petsko GA. Structure of yeast triosephosphate isomerase at 1.9-A resolution. Biochemistry. 1990 Jul 17;29(28):6609-18. PMID: 2204417 Pompliano DL, Peyman A, Knowles JR. Stabilization of a reaction intermediate as a catalytic device: definition of the functional role of the flexible loop in triosephosphate isomerase. Biochemistry. 1990 Apr 3;29(13):3186-94. PMID: 2185832
| ||||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Paula Grabowski, Jacqueline Townsend, Joel L. Sussman, Kara Pryke, Regina D. Kettering, Gregg Snider