AlphaFold2 examples from CASP 14
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
The quality of predictions for the structure of ORF8 are judged by comparison with X-ray crystallographic [[empirical models]] which were not available to the groups making predictions. Shortly after the CASP 14 competition (summer 2020), two X-ray crystal structures were reported for ORF8: [[7jtl]] released August 26, 2020, and [[7jx6]], released September 23, 2020. The [[resolution|resolutions]] are 2.0 and 1.6 Å respectively, and both have worse than average [[Rfree]] values. | The quality of predictions for the structure of ORF8 are judged by comparison with X-ray crystallographic [[empirical models]] which were not available to the groups making predictions. Shortly after the CASP 14 competition (summer 2020), two X-ray crystal structures were reported for ORF8: [[7jtl]] released August 26, 2020, and [[7jx6]], released September 23, 2020. The [[resolution|resolutions]] are 2.0 and 1.6 Å respectively, and both have worse than average [[Rfree]] values. | ||
{{Template:Green links zoom}} | {{Template:Green links zoom}} | ||
| - | <scene name='87/875686/Chain_a_of_7jx6/1'>Here is one chain of ORF8</scene> from the higher resolution X-ray structure, [[7jx6]]. These chains form [http://firstglance.jmol.org/fg.htm?mol=7jx6 disulfide-linked dimers], and the dimers form higher order multimers<ref name=" | + | <scene name='87/875686/Chain_a_of_7jx6/1'>Here is one chain of ORF8</scene> from the higher resolution X-ray structure, [[7jx6]]. These chains form [http://firstglance.jmol.org/fg.htm?mol=7jx6 disulfide-linked dimers], and the dimers form higher order multimers<ref name="7jtl" /> (not shown). Notice that the <span class="text-blue"><b>amino</b></span> and <span class="text-red"><b>carboxy</b></span> '''ends of the chain come together''' to form two parallel beta strands of a beta sheet. Also notice that there are '''3 disulfide bonds'''. An accurate prediction would include both of these features. |
<scene name='87/875686/Morf_lin_7jx6_imf_7jtl/3'>The two X-ray structures agree very well</scene><ref name="imf">Superposition by Swiss-PdbViewer's ''iterative magic fit''. This starts with a sequence alignment-guided structural superposition, and then superposes subsets of the structures to minimize the RMSD. Eight intermediate structures were generated by the [[Morphs#Linear_Morph_Server|Theis Morph Server]] by linear interpolation.</ref>. The only substantial disagreement is for a large surface loop, sequence range 48-57. See the Table I below for [https://en.wikipedia.org/wiki/Root-mean-square_deviation_of_atomic_positions RMSD] values. | <scene name='87/875686/Morf_lin_7jx6_imf_7jtl/3'>The two X-ray structures agree very well</scene><ref name="imf">Superposition by Swiss-PdbViewer's ''iterative magic fit''. This starts with a sequence alignment-guided structural superposition, and then superposes subsets of the structures to minimize the RMSD. Eight intermediate structures were generated by the [[Morphs#Linear_Morph_Server|Theis Morph Server]] by linear interpolation.</ref>. The only substantial disagreement is for a large surface loop, sequence range 48-57. See the Table I below for [https://en.wikipedia.org/wiki/Root-mean-square_deviation_of_atomic_positions RMSD] values. | ||
Revision as of 22:43, 1 March 2021
This page is under construction. Eric Martz 01:03, 22 February 2021 (UTC)
Prediction of protein structures from amino acid sequences, theoretical modeling, has been extremely challenging. In 2020, breakthrough success was achieved by AlphaFold2[1], a project of DeepMind. For an overview of this breakthrough, documented by the bi-annual prediction competition CASP, please see 2020: CASP 14. Below are illustrated some examples of predictions from that competition.
| |||||||||||
Contents |
ORF8 Sidechain Accuracy
AlphaFold2's predictions for sidechain positions seem fairly good, while sidechain positions in the 2nd best prediction seem poor. This conclusion is based on three types of observations:
- Table I gives RMSD values for all atoms, which is one indication of sidechain accuracy.
- Prediction of salt bridges and cation-pi interactions.
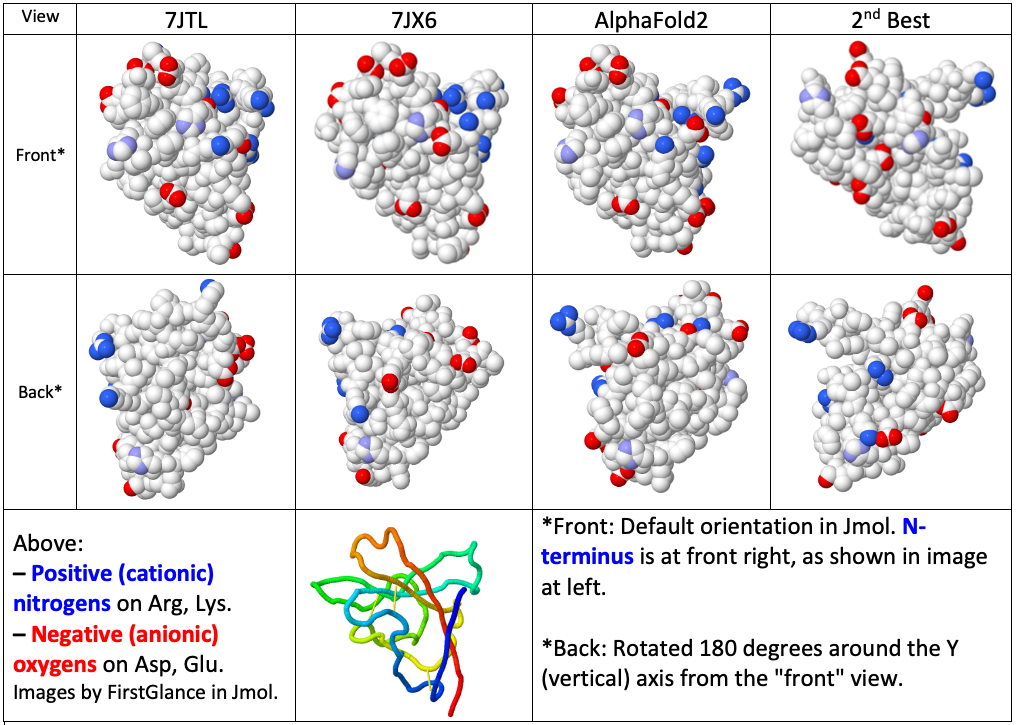
- Visualization of the distributions of charges on the surfaces.
Salt Bridges and Cation-Pi Interactions
- AlphaFold2's prediction was correct for 4/5 interactions, with one incorrect interaction.
- AlphaFold2's prediction was correct for one of two salt bridges, and predicted no incorrect salt bridges.
- AlphaFold2's prediction was correct for three of three cation-pi interactions, but predicted one incorrect interaction.
- The 2nd best prediction was correct for 1/5 interactions, with 2 incorrect interactions.
- The 2nd best prediction was correct for one of two salt bridges, but predicted two incorrect salt bridges.
- The 2nd best prediction failed to predict any of the three cation-pi interactions, predicting zero interactions.
| 7JX6 | 7JTL | AlphaFold2 | 2nd Best |
|---|---|---|---|
| R101:D112 (AB) | R101:D113 (AB) | R86:D98 | R86:D98 |
| R115:D119 (AB) | R115:D119 (AB) | – | R100:E4 |
| K44:E59 (AB) | K44:E59 (AB) | K29:E44 | – |
| – | – | – | K78:E77 |
- Bridges in the same row are identical (except for red residues). Subtract 15 from the sequence numbers in the X-ray structures for the equivalent sequence numbers in the predictions.
- Black: Shortest sidechain nitrogen to sidechain oxygen distance ≤4.0 Å.
- Gray: Shortest sidechain nitrogen to sidechain oxygen distance 4.4 to 4.8 Å.
- –: Shortest sidechain nitrogen to sidechain oxygen distance 6 to 16 Å.
- (AB): The two chains in each X-ray model.
- Italics: erroneous prediction.
| 7JX6 | 7JTL | AlphaFold2 | 2nd Best |
|---|---|---|---|
| R101:Y46+Y108 (AB) | R101:Y46+Y108 (AB) | R86:Y31+Y96 | – |
| K44:F108 (B) | K44:F108 (AB) | K29:F93 | – |
| – | – | K79:F105 | – |
- All interactions listed are deemed energetically significant by the CaPTURE Server.
- Interactions in the same row are identical. Subtract 15 from the sequence numbers in the X-ray structures for the equivalent sequence numbers in the predictions.
- Italics: erroneous prediction.
- The 2nd best prediction has no cation-pi interactions.
- (AB): The two chains in each X-ray model.
Visualization of Surface Charge Distributions
References
- ↑ Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Zidek A, Nelson AWR, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K, Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020 Jan;577(7792):706-710. doi: 10.1038/s41586-019-1923-7. Epub 2020 Jan, 15. PMID:31942072 doi:http://dx.doi.org/10.1038/s41586-019-1923-7
- ↑ CASP14: what Google DeepMind’s AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics, a blog post by Carlos Outeir al Rubiera, December 3, 2020.
- ↑ 3.0 3.1 Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. 2021 Jan 12;118(2). pii: 2021785118. doi:, 10.1073/pnas.2021785118. PMID:33361333 doi:http://dx.doi.org/10.1073/pnas.2021785118
- ↑ 4.0 4.1 Summary and Classifications of Domains for CASP 14.
- ↑ 5.0 5.1 5.2 5.3 Superposition by Swiss-PdbViewer's iterative magic fit. This starts with a sequence alignment-guided structural superposition, and then superposes subsets of the structures to minimize the RMSD. Eight intermediate structures were generated by the Theis Morph Server by linear interpolation.
- ↑ Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011 Jan;39(Database issue):D420-6. doi: 10.1093/nar/gkq1001. , Epub 2010 Nov 19. PMID:21097779 doi:http://dx.doi.org/10.1093/nar/gkq1001
- ↑ Holm L. DALI and the persistence of protein shape. Protein Sci. 2020 Jan;29(1):128-140. doi: 10.1002/pro.3749. Epub 2019 Nov 5. PMID:31606894 doi:http://dx.doi.org/10.1002/pro.3749
- ↑ Using Swiss-PdbViewer's Fit from Selection with 102 residues selected from each structure, followed by Improve Fit.
- ↑ Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013 Apr;30(4):772-80. doi: 10.1093/molbev/mst010. Epub 2013 Jan, 16. PMID:23329690 doi:http://dx.doi.org/10.1093/molbev/mst010
- ↑ Structural superposition by Dali. Interpolation by the Yale Morph2 Server. Homogenization method: homology modeling. No minimization. This produced a 9-model file where model 1 was 7jx6, and models 2-9 were interpolations. 5a2f residues 28-133 were added as model 10 (black in the molecular scene).
- ↑ The interpretation of Dali's result to mean that ORF8 does not have a novel fold was kindly confirmed by Liisa Holm, personal communication to Eric Martz.
- ↑ Download AlphaFold2's predicted structure for ORF8 from T1064TS427_1-D1.pdb.
- ↑ Superposition by Swiss-PdbViewer's magic fit. This is a sequence alignment-guided structural superposition. Eight intermediate structures were generated by the Theis Morph Server by linear interpolation.
- ↑ Superposition by Swiss-PdbViewer's Explore Fragment Alternate Fits, which does not use sequence information. Eight intermediate structures were generated by the Theis Morph Server by linear interpolation.
- ↑ For all targets in CASP 14, the top two servers were QUARK and Zhang-server (which were not significantly different at a Z-score sum of 62.9), followed by Zhang-CEthreader (55.9) and BAKER-ROSETTASERVER (55.3).