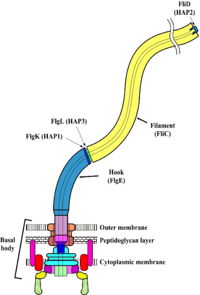
Flagellar protein
From Proteopedia
(Redirected from Flagella, bacterial)
| |||||||||||
References and Notes
- ↑ Lockman HA, Curtiss R 3rd. Salmonella typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect Immun. 1990 Jan;58(1):137-43. PMID:2152887
- ↑ Daniels R, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev. 2004 Jun;28(3):261-89. PMID:15449604
- ↑ Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009 Mar;17(3):109-18. Epub 2009 Feb 21. PMID:19231189 doi:10.1016/j.tim.2008.12.004
- ↑ Lemon KP, Higgins DE, Kolter R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol. 2007 Jun;189(12):4418-24. Epub 2007 Apr 6. PMID:17416647 doi:10.1128/JB.01967-06
- ↑ Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007 Oct;15(10):456-61. Epub 2007 Oct 24. PMID:17920274 doi:10.1016/j.tim.2007.09.006
- ↑ Matsunami H, Yoon YH, Meshcheryakov VA, Namba K, Samatey FA. Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica. Sci Rep. 2016 Jun 7;6:27399. doi: 10.1038/srep27399. PMID:27273476 doi:http://dx.doi.org/10.1038/srep27399
- ↑ Ohnishi K, Ohto Y, Aizawa S, Macnab RM, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994 Apr;176(8):2272-81. PMID:8157595
- ↑ You Y, Ye F, Mao W, Yang H, Lai J, Deng S. An overview of the structure and function of the flagellar hook FlgE protein. World J Microbiol Biotechnol. 2023 Mar 21;39(5):126. PMID:36941455 doi:10.1007/s11274-023-03568-6
- ↑ Saijo-Hamano Y, Uchida N, Namba K, Oosawa K. In vitro characterization of FlgB, FlgC, FlgF, FlgG, and FliE, flagellar basal body proteins of Salmonella. J Mol Biol. 2004 May 28;339(2):423-35. PMID:15136044 doi:10.1016/j.jmb.2004.03.070
- ↑ Herlihey FA, Moynihan PJ, Clarke AJ. The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J Biol Chem. 2014 Nov 7;289(45):31029-42. PMID:25248745 doi:10.1074/jbc.M114.603944
- ↑ Bulieris PV, Shaikh NH, Freddolino PL, Samatey FA. Structure of FlgK reveals the divergence of the bacterial Hook-Filament Junction of Campylobacter. Sci Rep. 2017 Nov 16;7(1):15743. doi: 10.1038/s41598-017-15837-0. PMID:29147015 doi:http://dx.doi.org/10.1038/s41598-017-15837-0
- ↑ Inoue Y, Kinoshita M, Kida M, Takekawa N, Namba K, Imada K, Minamino T. The FlhA linker mediates flagellar protein export switching during flagellar assembly. Commun Biol. 2021 May 31;4(1):646. PMID:34059784 doi:10.1038/s42003-021-02177-z
- ↑ Meshcheryakov VA, Barker CS, Kostyukova AS, Samatey FA. Function of FlhB, a membrane protein implicated in the bacterial flagellar type III secretion system. PLoS One. 2013 Jul 11;8(7):e68384. doi: 10.1371/journal.pone.0068384. Print 2013. PMID:23874605 doi:http://dx.doi.org/10.1371/journal.pone.0068384
- ↑ Lee J, Harshey RM. Loss of FlhE in the flagellar Type III secretion system allows proton influx into Salmonella and Escherichia coli. Mol Microbiol. 2012 May;84(3):550-65. PMID:22435757 doi:10.1111/j.1365-2958.2012.08043.x
- ↑ Zhang K, He J, Cantalano C, Guo Y, Liu J, Li C. FlhF regulates the number and configuration of periplasmic flagella in Borrelia burgdorferi. Mol Microbiol. 2020 Jun;113(6):1122-1139. PMID:32039533 doi:10.1111/mmi.14482
- ↑ Postel S, Deredge D, Bonsor DA, Yu X, Diederichs K, Helmsing S, Vromen A, Friedler A, Hust M, Egelman EH, Beckett D, Wintrode PL, Sundberg EJ. Bacterial flagellar capping proteins adopt diverse oligomeric states. Elife. 2016 Sep 24;5. pii: e18857. doi: 10.7554/eLife.18857. PMID:27664419 doi:http://dx.doi.org/10.7554/eLife.18857
- ↑ Ikeda T, Oosawa K, Hotani H. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol. 1996 Jun 21;259(4):679-86. PMID:8683574 doi:10.1006/jmbi.1996.0349
- ↑ Levenson R, Zhou H, Dahlquist FW. Structural insights into the interaction between the bacterial flagellar motor proteins FliF and FliG. Biochemistry. 2012 Jun 26;51(25):5052-60. PMID:22670715 doi:10.1021/bi3004582
- ↑ Minamino T, Kinoshita M, Inoue Y, Morimoto YV, Ihara K, Koya S, Hara N, Nishioka N, Kojima S, Homma M, Namba K. FliH and FliI ensure efficient energy coupling of flagellar type III protein export in Salmonella. Microbiologyopen. 2016 Jun;5(3):424-35. PMID:26916245 doi:10.1002/mbo3.340
- ↑ Tachiyama S, Chan KL, Liu X, Hathroubi S, Peterson B, Khan MF, Ottemann KM, Liu J, Roujeinikova A. The flagellar motor protein FliL forms a scaffold of circumferentially positioned rings required for stator activation. Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2118401119. PMID:35046042 doi:10.1073/pnas.2118401119
- ↑ Toker AS, Macnab RM. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997 Oct 31;273(3):623-34. PMID:9356251 doi:10.1006/jmbi.1997.1335
- ↑ Paul K, Blair DF. Organization of FliN subunits in the flagellar motor of Escherichia coli. J Bacteriol. 2006 Apr;188(7):2502-11. PMID:16547037 doi:10.1128/JB.188.7.2502-2511.2006
- ↑ Ward E, Renault TT, Kim EA, Erhardt M, Hughes KT, Blair DF. Type-III secretion pore formed by flagellar protein FliP. Mol Microbiol. 2018 Jan;107(1):94-103. PMID:29076571 doi:10.1111/mmi.13870
- ↑ Ozin AJ, Claret L, Auvray F, Hughes C. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS Microbiol Lett. 2003 Feb 28;219(2):219-24. PMID:12620624 doi:10.1016/S0378-1097(02)01208-9
- ↑ Minamino T, Kinoshita M, Imada K, Namba K. Interaction between FliI ATPase and a flagellar chaperone FliT during bacterial flagellar protein export. Mol Microbiol. 2012 Jan;83(1):168-78. PMID:22111876 doi:10.1111/j.1365-2958.2011.07924.x
- ↑ Bischoff DS, Ordal GW. Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol Microbiol. 1992 Sep;6(18):2715-23. PMID:1447979 doi:10.1111/j.1365-2958.1992.tb01448.x
- ↑ Wunder EA Jr, Figueira CP, Benaroudj N, Hu B, Tong BA, Trajtenberg F, Liu J, Reis MG, Charon NW, Buschiazzo A, Picardeau M, Ko AI. A novel flagellar sheath protein, FcpA, determines filament coiling, translational motility and virulence for the Leptospira spirochete. Mol Microbiol. 2016 Aug;101(3):457-70. PMID:27113476 doi:10.1111/mmi.13403
- ↑ Wunder EA Jr, Slamti L, Suwondo DN, Gibson KH, Shang Z, Sindelar CV, Trajtenberg F, Buschiazzo A, Ko AI, Picardeau M. FcpB Is a Surface Filament Protein of the Endoflagellum Required for the Motility of the Spirochete Leptospira. Front Cell Infect Microbiol. 2018 May 8;8:130. PMID:29868490 doi:10.3389/fcimb.2018.00130
- ↑ Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol Biosyst. 2008 Nov;4(11):1105-15. Epub 2008 Sep 24. PMID:18931786 doi:10.1039/b808065h
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Eric Martz, Joel L. Sussman, Fadel A. Samatey, Jaime Prilusky