User:Fadel A. Samatey/FlgE III/Intrinsically Disordered Flagellar Rod Stretch
From Proteopedia
Interactive 3D Complement in Proteopedia
BMC Biology an online-only, open access journal: bmcbiol
An intrinsically disordered linker controlling the formation and the stability of the bacterial flagellar hook.
Clive S. Barker, Irina V. Meshcheryakova, Alla S. Kostyukova, Peter L. Freddolino, and Fadel A. Samatey.
BMC Biology 15:97 (October 27, 2017) (doi.org/10.1186/s12915-017-0438-7)
The interactive Molecular Tour below assumes that you are familiar with the journal article[1].
|
In addition to empirical structures, this report includes some theoretical models, which should be treated with caution. |
Contents |
Introduction
|
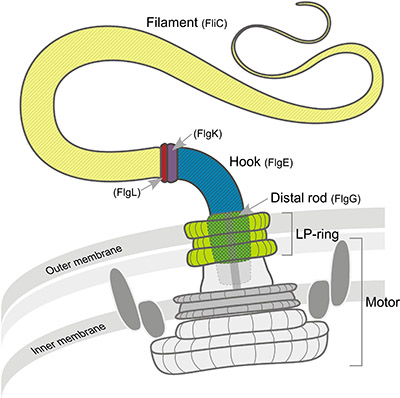
The flagellar hook is the universal joint that transmits torque from the motor, via the flagellar rod, to the helical flagellar filament which propels motile bacteria. Crystallographic structures of the hook monomer protein[2][3] and a cryo-EM structure of the rod monomer protein[4] have been reported. Flagellar hook and rod proteins have a segment that appears likely to be intrinsically disordered before assembly of the hook or rod. For reasons explained in the publication[1], we shall refer to this segment as the Intrinsically Disordered Rod Stretch, ID-Rod-Stretch (in both the hook and the rod). "Rod" is included in the name because this segment is more conserved in the rod (FlgG)[1]. A less conserved homolog occurs in the hook (FlgE). This segment is missing in several earlier hook and rod protein monomer structures[2][5][3][4]. Previously, we reported a complete structure of the Campylobacter jejuni hook by cryo-EM at 3.5 Å[6]. This provided the first structure of the ID-Rod-Stretch. We showed that after assembly of the hook, the ID-Rod-Stretch (earlier termed the L-stretch) contacts several adjacent monomer proteins, assuming a stable folded structure. Evidence presented in the current report[1] demonstrates that the ID-Rod-Stretch is crucial to stabilization and strength of the hook and rod. Because of its dual nature, we refer to the ID-Rod-Stretch as the yin and yang of the flagellum. (Please scroll down to continue) | |
|
The bacterial flagellum consists of a filament, a universal joint ("hook"), and a motor ("basal body") containing a "rod" that transmits torque to the hook. This is Figure 1 in [1]. |
Molecular Tour
| |||||||||||
See Also
- YouTube video about this work.
- Complete 3.5Å cryo-EM structure of the Campylobacter flagellar hook.
- The Bacterial Flagellar Hook
- Flagella, bacterial
Notes and References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Barker CS, Meshcheryakova IV, Kostyukova AS, Freddolino PL, Samatey FA. An intrinsically disordered linker controlling the formation and the stability of the bacterial flagellar hook. BMC Biol. 2017 Oct 27;15(1):97. doi: 10.1186/s12915-017-0438-7. PMID:29078764 doi:http://dx.doi.org/10.1186/s12915-017-0438-7
- ↑ 2.0 2.1 2.2 Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, Derosier DJ, Kitao A, Namba K. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 2004 Oct 28;431(7012):1062-8. PMID:15510139 doi:http://dx.doi.org/10.1038/nature02997
- ↑ 3.0 3.1 3.2 Yoon YH, Barker CS, Bulieris PV, Matsunami H, Samatey FA. Structural insights into bacterial flagellar hooks similarities and specificities. Sci Rep. 2016 Oct 19;6:35552. doi: 10.1038/srep35552. PMID:27759043 doi:http://dx.doi.org/10.1038/srep35552
- ↑ 4.0 4.1 4.2 Fujii T, Kato T, Hiraoka KD, Miyata T, Minamino T, Chevance FF, Hughes KT, Namba K. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun. 2017 Jan 25;8:14276. doi: 10.1038/ncomms14276. PMID:28120828 doi:http://dx.doi.org/10.1038/ncomms14276
- ↑ Shaikh TR, Thomas DR, Chen JZ, Samatey FA, Matsunami H, Imada K, Namba K, Derosier DJ. A partial atomic structure for the flagellar hook of Salmonella typhimurium. Proc Natl Acad Sci U S A. 2005 Jan 25;102(4):1023-8. Epub 2005 Jan 18. PMID:15657146
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Matsunami H, Barker CS, Yoon YH, Wolf M, Samatey FA. Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat Commun. 2016 Nov 4;7:13425. doi: 10.1038/ncomms13425. PMID:27811912 doi:http://dx.doi.org/10.1038/ncomms13425
- ↑ 7.0 7.1 "Contacting" is defined as likely hydrogen bonds, plus likely apolar interactions. Likely hydrogen bonds: oxygens or nitrogens within 3.5 Å of oxygens or nitrogens in a neighboring monomer. Apolar interactions: carbons or sulfurs within 4.0 Å of carbons or sulfurs in a neighboring monomer.
- ↑ Salt bridges to the ID-Rod-Stretch: One ID-Rod-Stretch Arg58 sidechain nitrogen is 2.3 Å from the closest oxygen of Glu802, and 1.8 Å from the closest oxygen of Asp105. The Glu and Asp are in different chains.
- ↑ Cation-pi interaction with the ID-Rod-Stretch: Phe133 is close enough to Arg58 in the ID-Rod-Stretch that an interaction is possible. In the cryo-EM model, the Phe133 ring is in the wrong orientation to make this energetically significant, but such an interaction may occur given the uncertainties in that model.
- ↑ The model Image:Hkcj d10.pdb.gz contains one chain "d" from the 55-monomer cryo-EM hook model, plus all atoms within 10 Å of that chain. Chain "d" was verified to have 268 contacting atoms, the maximum number present for any of the 55 chains. This "chain d + 10 Å" model was uploaded into FirstGlance in Jmol. There, in the Tools tab, the tool Contacts and Non-Covalent Interactions was employed to analyze the types of non-covalent bonds to the ID-Rod-Stretch.
- ↑ 11.0 11.1 11.2 Clashscores were determined by Molprobity as the number of serious steric atomic overlaps (> 0.4 Å) per 1000 atoms, after addition and optimization of hydrogen atoms. Zero percentile is worst (highest clashscore) and 100th percentile is the best, based on 1,724 reference structures chosen in 2004.
- ↑ 12.0 12.1 Sequence numbers start with 1 at the second residue in the genomic sequence, since the initial Met is believed to be removed by N-terminal methionine aminopeptidase.
- ↑ 13.0 13.1 The model of the Salmonella enterica hook was constructed using the 7.1 Å cryo-EM monomer 3a69 plus the ID-Rod-Stretch homology modeled using the Campylobacter jejuni 3.5 Å cryo-EM monomer 5jxl as template. 3a69 was constructed by adjusting the interdomain angles of the 1.8 Å X-ray structure of domains 1 and 2, 1wlg, for optimal fit into the cryo-EM map.
- ↑ UniProt sequence P0A1J3 (FLGG_SALTY) was submitted to Swiss Model, with the template specified as 5jxl. Two homology models were returned, one spanning residues 2-128 (42.7% sequence identity), and one spanning residues 175-260 (41.9% sequence identity). When combined, these provided a model of D0, and a partial model of D1. The monomer model was completed using the 7.4 Å cryo-EM model 5wrh.
- ↑ Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014 Jul;42(Web Server issue):W252-8. doi: 10.1093/nar/gku340., Epub 2014 Apr 29. PMID:24782522 doi:http://dx.doi.org/10.1093/nar/gku340
- ↑ Helical symmetry parameters for Salmonella enterica: hook rotation 64.79 degrees, rise 4.12 Å; rod rotation 64.72 degrees, rise 4.13 Å.
- ↑ Fujii T, Kato T, Namba K. Specific arrangement of alpha-helical coiled coils in the core domain of the bacterial flagellar hook for the universal joint function. Structure. 2009 Nov 11;17(11):1485-93. PMID:19913483 doi:10.1016/j.str.2009.08.017