Function
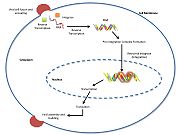
Retroviral Integrase is an essential retroviral enzyme that binds to viral DNA and inserts it into a host cell chromosome. The complex of integrase and DNA is called intasome. The reverse transcribed cDNA of human immunodeficiency virus type 1 (HIV-1) is inserted in the host cell genome in order increase pathogen fitness and virulence. Integrase is produced by a class of retrovirus (like HIV) and is used by the virus to incorporate its genetic material into the host cell DNA. The host cellular machinery then produces mRNA and then protein from the incorporated genetic material, thus replicating the virus. Although several integrase inhibiting drugs have been investigated, the mechanism responsible for strand-transfer inhibition action remains to be elucidated [1]. However, Hare el al (2010)[2] determined the structural constituents of retroviral integration. Further elucidation of the complete structure of the retroviral integrase, and its application to regulate functional and enzymatic activities could potentially enable researchers to delay the progression of retroviral diseases. Moreover, study of HIV-1 integration could lead to a promising new target, and contribute to the generation pharmacophore models for antiviral therapy[3].
HIV Integrase inhibitors: Raltegravir, marketed as Isentress is currently approved as a therapeutic inhibitor of HIV integrase. It was approved on October 12, 2007.
[See below for a table of antiretroviral drugs with trade name, company, patents, and notes.]
For phage integrase see Phage integrase.
Mechanism for strand-transfer inhibition actio
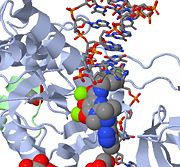
Above is a JMOL image of MK-0518 blocking the 3' end of the viral DNA from binding the active site. Notice the metal chelating oxygen atoms in MK-0518 interacting with the magnesium cations found in the active site.
HIV integrase type 1 is a relatively new and novel target for inhibitors. In 2007, the first HIV-1 integrase inhibitor, Raltegravir, was approved by the FDA for use in HIV-1 as a combination therapy [4]. Strand-transfer inhibitors work by preventing the concerted integration of the viral DNA into the host chromosome. After viral entry into the host cell, the reverse transcriptase converts the viral ssRNA into dsDNA. At this point, the integrase forms a complex with the viral DNA, creating the pre-integration complex (intasome). The pre-integration complex is then chaperoned into the nucleus, where two nucleotides are excised from the 3' end. Next, the DNA is then covalently integrated into the host DNA. The strand-transfer inhibitors interrupt this process, preventing the integration of the viral DNA into the host chromosome. Strand-transfer inhibitors work by engaging the metal ion cofactors found in the retroviral integrase active site. The metal chelating oxygen atoms found in the inhibitors interact directly with the metal cofactors, while the halobenzyl group fits into the pocket created by the displaced 3' viral DNA in the active site.
HIV and AIDS

In 2010, there are more than 25 million people have died of AIDS, and it is estimated that approximately 33 million people are living with HIV.
To date, more than 25 million people have died of AIDS and it is estimated that approximately 33 million people are living with HIV today[5]. For retroviral integrase inhibitors see Raltegravir, Genvoya and Retroviral Integrase Inhibitor Pharmacokinetics.
Impact of Structure
Three-dimensional structures for certain host-cell proteins critical to understanding the mechanism of HIV infection and virulence have emerged from X-ray crystallographic analyses. HIV protease and integrase structures are among the highest ranked structures that have contributed to saving many lives and added to the quality of life of many HIV-afflicted individuals. It is implemented in structure-based drug design to develop protease inhibitors and integrase inhibitors, and is used as a significant component of highly active anti-retroviral therapy (HAART).
While existing antiretroviral agents improve the quality of life as well as extending the life of many patients, it fails to eradicate the disease. Studies in integrase inhibitors show that combination with other antiretroviral drugs diminish viral adaptations, and may have the potential to be used for salvage therapy for patients who have acquired resistance to other drugs. For more, please see
PFV Intasome Crystallization
To mimic the viral DNA ends of HIV-1, Hare et al (2010)[2] utilized soluble and fully functional prototype foamy virus (PFV) intasome preparations, obtained using recombinant PFV integrase and double-stranded oligonucleotides.
The remarkable stability of the integrase-DNA complexes were determined by observing the in vitro strand transfer reactions, which were classified into three modes of deproteination migration: (1) single concerted events: linearized target plasmid; (2) multiple concerted events: smear; (3) half-site events: open circular DNA. Further characterization of the PFV intasome also exhibited structural substantiality which implied strong protein-protein and protein-DNA interactions despite prolonged incubation under high ionic strength conditions. Comprehensive crystallization assays effected a viable crystal configuration that diffracted X-rays to 2.9 Angstroms resolution. A three-dimensional structure was ultimately determined. The asymmetric unit contained a single integrase dimer with a stably bound viral DNA molecule, and a pair of integrase dimers consociated with symmetry, which formed an oblong tetramer. The dimer interface is stabilized by intermolecular amino terminal and catalytic core domains (inner subunit-outer subunit) interactions. The overall shape of the oblong tetramer is unique albeit bearing semblances to previously reported HIV-1 integrase complexes.
Crystallization Technique
The protein-DNA complexes were formed using the full-length, wild-type PFV IN and synthetic dsDNA that modeled the viral end.
The intasome was crystallized utilizing the vapor-drop hanging diffusion technique. The reservoir solution consisted on 1.35 M ammonium sulphate, 25% (v/v) glycerol, 4.8% (v/v) 1,6-hexanediol, and 50mM 2-(N-morpholino) ethanesulphonic acid (MES) at pH 6.5. The protein-DNA crystals were also soaked in the presence of MK0518, GS9137, Mg(II), and/or Mn(II). The crystal structure was solved using molecular replacement.
Crystallographic and Refinement Statistics
Hare et al (2010)[2] have published data on seven crystal structures. These data include the PFV IN complex (apo form) and six additional structures, including the complex bound to Mg, Mn, Mg/MK0518, Mn/MK0518, Mg/GS9137, and Mn/GS9137. All seven structures belong to the P41212 space group. They have been refined to between 2.85 and 3.25 Å resolution.
Overall Architecture & Components
Structure
The overall structure of the assembled PFV intasome is a tetramer model based on two domain structures with a dimer-dimer interface. Previous intasome models depict a similar but more flexible structure while the PFV intasome has been shown to be highly constrained. Using homology modeling, Hare et al (2010)[2] propose that shorter interdomain linkers may be a factor in flexibility, specifically in HIV-1 integrase. The inner subunits of the tetramer are implicated in the overall tetramerization and viral DNA binding. The catalytic core domains of the outer subunits may act as supports, but since the amino- and carboxy-terminal domains are unresolved in electron density maps, their function remains inconclusive. The catalytic core domain and carboxy-terminal domain linker adopts an extended conformation for most of its length, and are located parallel to the amino-terminal domain and catalytic core domain linker of the inner subunit. The interdomain linkers The interdomain linkers (CCD-CTD linker and NTD-CCD linker) bind both halves of the intasome together, and the structure is further stabilized by a pair of carboxy-terminal domains interacting with both inner catalytic core domains.
Integrase and DNA interactions
Strong protein-DNA interactions are located within the terminal six nucleotides. Each carboxy terminal domain interacts with the phosphodiester backbone of both viral DNA molecules. Furthermore, the amino-terminal domain-extension domain and amino terminal domain interact with the viral DNA at the active site of the opposing catalytic core domain.
Active Site
Active site carboxylates are side chains of Asp 128, Asp 185, Glu 221.
One zinc atom each is located near the the active sites.
Integrase Inhibitors
| Name | Brand | Company | Patent | Notes
|
| Raltegravir | Isentress | Merck & Co. | - | also known as MK-0518. The isopropyl and methyl-oxadiazole of MK-0518 are involved in hydrophobic and stacking interactions with side chains of Pro 214 and Tyr 212 to stabilize this drug within the PFV intasome active site. This manner of drug-binding interaction causes displacement of the reactive 3' viral DNA end from the active site of PFV intasome. After binding of MK-0518 to active site, the reactive 3' hydroxyl group moves away from the active site of the PFV intasome by more than 6 Angstroms. Raltegravir was approved by the FDA on October 12, 2007, for use with other anti-HIV agents in the treatment of HIV infection in adults. It is the first integrase inhibitor approved by the FDA.
|
| Elvitegravir | - | Gilead Science | - | GS-9137 interacts with Pro 214 of PFV intasome through its quinolone base and isopropyl group. In experimental stages; shares the core structure of quinolone antibiotics. Phase II studies of elvitegravir in people who are treatment experienced have been completed. Phase III studies in treatment experienced patients are ongoing. A phase II study of elvitegravir in people who have never taken antiretroviral therapy is underway. This study will also be evaluated a boosting agent in place of Norvir, currently called GS9350. Elvitegravir holds promise for HIV-positive patients who have taken other anti-HIV drugs in the past.
|
| MK-2048 | - | Merck & Co. | - | A second generation integrase inhibitor, intended to be used against HIV infection. It is superior to the first available integrase inhibitor, raltegravir, in that it inhibits the HIV enzyme integrase 4 times longer. It is being investigated for use as part of pre-exposure prophylaxis (PrEP).
|
See also Retroviral Integrase Inhibitor Pharmacokinetics[6].
Additional Resources
For additional information, see: Human Immunodeficiency Virus
3D structures of retroviral integrase
Retroviral integrase 3D structures