We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
SelB
From Proteopedia
| |||||||||||
3D structures of SelB
1lva – MtSelB C terminal – Moorella thermoacetica
2v9v - MtSelB winged-helix domains
4aca – MmSelB – Methanococcus maripaludis
SelB complex with nucleotides
4ac9 – MmSelB + GDP
4acb – MmSelB + GPPNHP
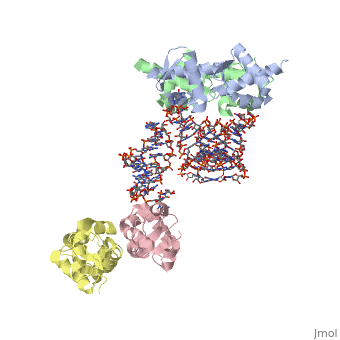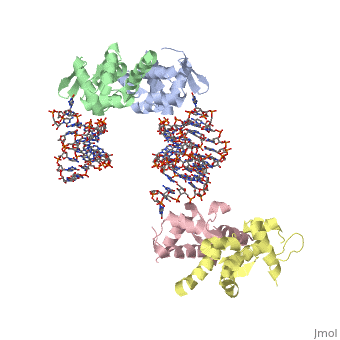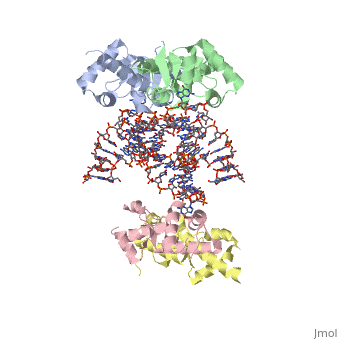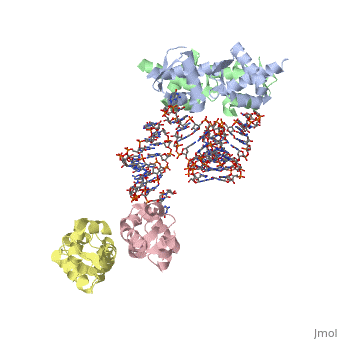
1wsu, 2uwm - MtSelB C terminal + RNA
2pjp – EcSelB mRNA-binding domain + RNA – Eschrichia coli
2ply - EcSelB mRNA-binding domain (mutant) + RNA
Additional Resources
For additional information, see: Translation
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Eran Hodis, David S. Goodsell, Joel L. Sussman, David Canner, Eric Martz, Alexander Berchansky