Electron density maps
From Proteopedia
(→Why Look At Electron Density Maps?) |
(→Outside of Proteopedia) |
||
| Line 52: | Line 52: | ||
*[http://en.wikipedia.org/wiki/Electron_density Electron density map at Wikipedia]. | *[http://en.wikipedia.org/wiki/Electron_density Electron density map at Wikipedia]. | ||
*[http://www.bioinformatics.org/molvis/edm/ Electron Density Maps in Jmol]. | *[http://www.bioinformatics.org/molvis/edm/ Electron Density Maps in Jmol]. | ||
| + | |||
| + | ==References== | ||
| + | <references /> | ||
Revision as of 19:41, 25 July 2021
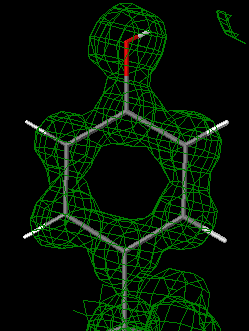
85% of the macromolecular structures available from the Protein Data Bank (PDB) were determined by X-ray crystallography. The direct results of crystallographic experiments are electron density maps. Examining the correspondence between the electron density map and the published molecular model reveals the levels of uncertainty in the model.
Contents |
Crystallography Produces Electron Density Maps
An X-ray crystallographic experiment produces an electron density map for the average unit cell of the protein crystal. The amino acid (or nucleotide) sequence of the crystallized polymer(s) is known in advance. The crystallographer fits the atoms of the known molecules into the electron density map, and refines the model and map to the limits of the resolution of the crystal (which is limited by the level of order or disorder in the crystal). The crystallographer then deposits a model of the asymmetric unit of the crystal in the PDB, along with the experimental diffraction data (amplitudes and widths of the X-ray reflection spots, or "structure factors") from which the electron density map can be reconstructed. Electron density maps are available for most PDB files from the Uppsala Electron Density Map Server.
Why Look At Electron Density Maps?
Examining the correspondence between the published model PDB file and the electron density map (EDM) provides much clearer insight into the uncertainties in the model than does merely examining the model itself[1] (see also Quality assessment for molecular models). In addition to examining the entire map (2mFo-DFc) it is revealing to examine the difference map (mFo-DFc), which shows where the model fails to account for the map.
Visualizing Electron Density Maps
Crystallographers generally use "heavy duty" visualization and modeling software such as Coot or PyMOL, which require considerable practice to use effectively. Jmol first became capable of displaying electron density maps in January, 2010. Being able to display EDM's in Jmol opens the door to examining EDMs effectively in a web browser, with a user interface (yet to be developed) that requires no specialized software knowledge.
Examples
In Proteopedia
- User:Karsten Theis/Electron density explains and illustrates how to display the electron density map for any selected portion of a crystallographic structure, at various sigma levels.
- Some of the green links in Garman lab: Interconversion of lysosomal enzyme specificities show electron density maps for ligands.
Outside of Proteopedia
- Electron Density: Cloud vs. Isomesh "Map" shows a "raw" electron density map with buttons to hide densities below various sigma "noise" levels. It also shows the isomesh at 1.0 sigma, and the atomic model fitted to the isomesh.
- Electron Density Maps shows the map for bacterial flagellin, resolution 2.0 Å, and shows how the temperature/B-factor relates to the map in areas of low and high temperature.
See Also
Within Proteopedia
- Hydrogen in macromolecular models shows an example of hydrogens visible in an electron density map at 0.69 Å resolution.
- X-ray crystallography
- Resolution
Outside of Proteopedia
References
- ↑ Lamb AL, Kappock TJ, Silvaggi NR. You are lost without a map: Navigating the sea of protein structures. Biochim Biophys Acta. 2015 Apr;1854(4):258-68. doi: 10.1016/j.bbapap.2014.12.021., Epub 2014 Dec 29. PMID:25554228 doi:http://dx.doi.org/10.1016/j.bbapap.2014.12.021
