COVID-19 AlphaFold2 Models
From Proteopedia
Background
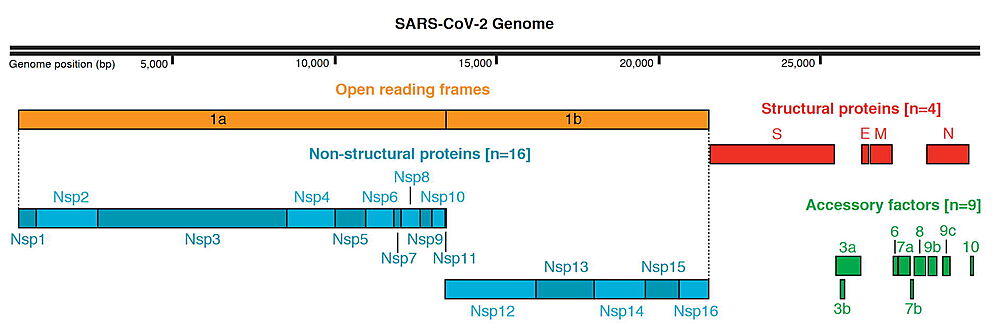
At present, there are still a number of proteins from the SARS CoV-2 virus whose 3D structures have not yet been experimentally determined. AlphaFold2 was used to predict these structures using the MIT ColabFold server[3], which was developed by Sergey Ovchinnikov, Milot Mirdita and Martin Steinegger. For each prediction, five 3D models were predicted, ranked from 1 to 5 (with 1 being the best). Views of these AlphaFold2 predictions can be seen on the Proteopedia pages:
AlphaFold2 Predicted Structures
| The quality of SARS-CoV-2 experimentally determined structures varies widely (Grabowski et al., 2021). Validated and corrected structures can be obtained from COVID19.BioReproducibility.Org. |
SARS-CoV-2 protein NSP6 - AlphaFold2 Theoretical Model - Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic[3][4][5].
SARS-CoV-2_protein_NSP11 - AlphaFold2 Theoretical Model - is the 13 amino acid peptide at the C-term of the large SARS-CoV-2 polyprotein ORF1ab (pp1a). Neither its functions, nor if it is expressed in vivo is clear[3][4][5]..
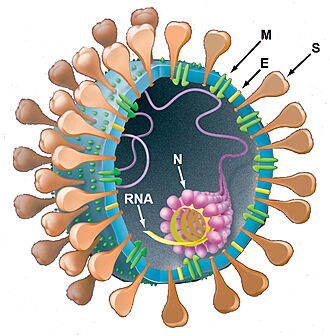
SARS-CoV-2 protein M - AlphaFold2 Theoretical Model - Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins[3][4][5].
SARS-CoV-2_protein_ORF6 - AlphaFold2 Theoretical Model : Could be a determinant of virus virulence[3][4][5].
SARS-CoV-2 protein N - AlphaFold2 Theoretical Model - The primary function of the Nucleocapsid protein (N-protein) is to package the viral genome into a helical ribonucleoprotein (RNP) complex[3][4][5].
SARS-CoV-2 protein ORF10 - AlphaFold2 Theoretical Model - It is currently unclear whether this region translates into a functional protein[3][4][5].
See also
Coronavirus_Disease 2019 (COVID-19)
SARS-CoV-2_virus_proteins
References
- ↑ Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Huttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O'Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d'Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, Garcia-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 Jul;583(7816):459-468. doi: 10.1038/s41586-020-2286-9. Epub 2020 Apr, 30. PMID:32353859 doi:http://dx.doi.org/10.1038/s41586-020-2286-9
- ↑ Holmes KV, Enjuanes L. Virology. The SARS coronavirus: a postgenomic era. Science. 2003 May 30;300(5624):1377-8. doi: 10.1126/science.1086418. PMID:12775826 doi:http://dx.doi.org/10.1126/science.1086418
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 MIT ColabFold
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Modeling of the SARS-COV-2 Genome
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J Proteome Res. 2020 Apr 3;19(4):1351-1360. doi: 10.1021/acs.jproteome.0c00129., Epub 2020 Mar 24. PMID:32200634 doi:http://dx.doi.org/10.1021/acs.jproteome.0c00129
