Function
Mechanosensitive channels are involved in touch, hearing, and in maintaining osmotic balance. In prokaryotes, they are opened by osmotic stretching of the lipid bilayer in which they are embedded. In eukaroytes, gating involves the cytoskeleton[1]. Mechanosensitive channels protect bacteria from sudden exposure to fresh water (water with lower osmolarity). Such exposure results in swelling of the bacterial cells due to osmotically-driven water influx. This swelling stretches the lipid bilayers, opening the mechanosensitive channels that are embedded in the inner membrane. The open channels allow rapid release of osmotically active ions or small molecules from the cytoplasm, improving the osmotic balance between the inside and outside of the cells, and preventing rupturing of the cell.
Models Available
Models are available for the open and closed forms of two mechanosensitive channels. These are illustrated below along with the potassium ion channel (which is not mechanosensitive) for comparison.
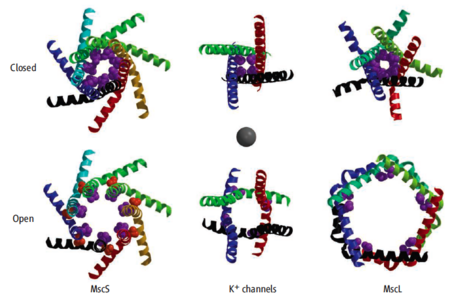
Closed and open forms of three gated channels, viewed from the outside of a cell. Only the innermost alpha helices that form the channel are shown. Sources of coordinates are in the table below. Magenta atoms are in hydrophobic residues that constrict the closed pores. Orange atoms in open MscS are A106V. Gray sphere in the center is 9 Å in diameter, approximating the threshold between water conducting and non-conducting hydrophobic pores. Figure from Gandhi and Rees,
Science 321:1166, 2008
[2] reproduced here with permission from AAAS and from Douglas Rees.
| Channel
| Closed
| Open
|
| MscS[3]
| 2oau, E. coli 3.7 Å 2002
| 2vv5, E. coli 3.5 Å 2008
|
| K+ Channels
| 1k4c, Streptomyces lividans 2.0 Å 2001
| 2r9r, Rattus norvegicus 2.4 Å 2007
|
| MscL[4]
| 2oar, Mycobacterium tuberculosis 3.5 Å 1998/2006
1kyk, 1kyl, Theoretical homology models, E. coli 2002
| 1kym, Theoretical model, E. coli 2002
|
Although there is agreement that 2oau represents a closed conformation, it may not be the initial closed conformation. Vásquez et al.[1] propose that "Except for the narrowing at the intracellular end of its pore, 2oau could, in principle, support ion conduction and thus might represent an inactivated/desensitized conformation after opening." A similar proposal was made earlier by Anishkin and Sukharev[5].
Theoretical models of closed, intermediate, and open forms of MscL may be seen at 2oar.
Structure of MscS
E. coli MscS[3] consists of seven identical chains (a homoheptamer). In the
here, each chain is given a distinct pastel color.
show the backbone traces, which can alternatively
be
as
Alpha Helices or Beta Strands .
The transmembrane end of the molecule has a predominantly
Hydrophobic
surface, while the surface of the cytoplasmic domain includes many
Polar
hydrophilic amino acids.

. The distribution of hydrophobic surface residues allows the
positions of the surfaces of the lipid bilayer
to be predicted[6].
. However, the predicted bilayer (24 Å[6]) is too thin because the crystallographic model lacks the N-terminal 46 residues[7] which include a short hydrophobic stretch, and a HIS tag.
| Amino Terminus |
|
|
|
|
|
|
|
Carboxy Terminus |
It appears that the .
The channel is formed by the . There is a , but because this is hydrophobic, it is believed to block passage of hydrophilic solutes (see below).
[8] in a cut-away view (3 of the 7 chains are hidden) shows conservation around the narrow part of the channel, and around the openings in the cytoplasmic domain that are believed to provide access by ions and solutes to the channel. In particular, Leu105 has conservation level 8, and Leu109, level 9 (highest conservation; see below for significance).
Opening and Closing
If the animation does not start when the molecule appears, click the green link a second time. You may need to scroll up and down to start the animation[9].
The publication in August, 2008 of a crystallographic structure for an open form of E. coli MscS (mutant A106V; 2vv5)[3][10][1], together with the closed conformation reported in 2002 (2oau) illustrated above, made it straightforward to generate a morph of MscS[3] opening and closing[11]. In early 2009, these represent the only crystal structures available for the same gated channel protein in both open and closed conformations. (Other open/closed pairs are for different proteins.)
. When the entire molecule is morphed from the closed to open conformation, it is clear that the transmembrane helices tilt and slide against each other, while there is relatively little change in the cytoplasmic domain. (The apparent small rotation of the cytoplasmic domain is an artefact of the morphing method.)
Here is the same animation but beware: only the alpha carbon atoms are shown (at 3x their van der Waals diameter). Without sidechains, the size of the opening in the channel is not accurate (see below).
Here are shown.
, as shown in this scene, starting at the N-terminus, as TM1, TM2, and TM3, with TM3 being bent into TM3a and TM3b. TM3a forms the inner lining of the channel, and (white dots).
. For this morph[11], only the N-terminal helices are shown (residues 27-112: TM1, TM2, and TM3a). The hydrophobic portion of the channel is facing you (see below).
. This animated morph shows all non-hydrogen atoms. Here, the widening and narrowing of the passage in the channel can be clearly seen. The pore is about 5 Å in diameter in the "closed" conformation, and about 13 Å in the open conformation[10]. Although an opening remains in the "closed" conformation, computational studies indicate that pores need not be entirely closed to be non-conducting. Hydrophobic pores <9 Å or <13 Å in diameter have been predicted not to conduct water or ions respectively[12].
Here is the same morph .
Hydrophobic - Polar.
The sidechains of Leu 105 and Leu109 form a ring lining the narrowest point of the closed channel, making it hydrophobic and non-conductive.
This is colored the same as in the previous scene, but TM1 and TM2 have been hidden. Rotate this animation to view it from the side (channel vertical), and notice that in the open conformation, the TM3a helices are separated, while in the closed conformation, they are in lateral contact.
Here is a into the channel (with the front three chains hidden, leaving only the four in the back).
, viewed from inside the channel. A conserved pattern of Alanines and Glycines is said to form "knobs and holes" in the inter-chain TM3a contacts of the closed conformation[10].
.
Interchain Contacts
Although the changes in interchain contacts during opening and closing are of interest, the available models have modest resolution (3.5-3.7 Å) that is insufficient for sidechain positions to be known accurately. In addition, each of the seven chains in 2oau was apparently solved independently: when chains B-G are aligned[13], one at a time, with chain A, the median RMS deviation for alpha carbons is 1.54 Å (range 0.57-1.84). This could lead to variations in interchain contacts that may not be meaningful. The seven chains in 2vv5 were apparently restrained to similar conformations: the median alpha carbon RMS (between chain A and the other six) is only 0.07 Å (range 0.10 - 0.09 Å; sidechain RMS runs about 0.4 Å).
Chains B-G aligned to chain A for 2oau. (.)
for 2vv5.
Examination of the contacts to each of the seven chains in 2oau and in 2vv5 using FirstGlance in Jmol (details not shown) showed that the interchain interactions in the transmembrane region (as indicated by the predicted bilayer position, see above) are entirely hydrophobic (carbon-carbon), with the exception of one putative hydrogen bond between chains C and D (Arg88:C.NH2 is 2.9 Å from Val89:D.O) in 2oau. Thus, during opening and closing, the transmembrane helices could slide against each others' "oily" surfaces.
3D structures of mechanosensitive channels
Ion channels
See Also



