Mini Map Aide
From Proteopedia
The following article was written by Jonathan Cooper, Professor Emeritus at University College, London.
(Eric Martz put it into Proteopedia, but had no authorship role.) |
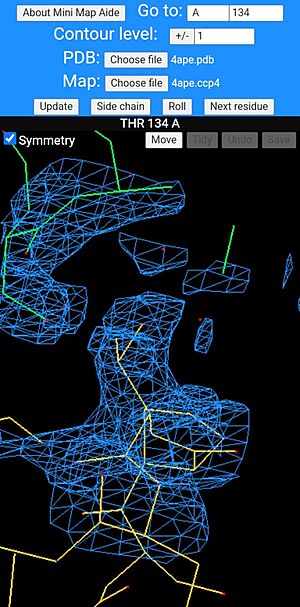
Mini map aide in use showing a threonine residue at the centre of the screen with a symmetry-related molecule, shown with its bonds drawn green. The bonds of main molecule are coloured dark yellow and individual atoms are coloured according to type, red for oxygen and blue for nitrogen.
Mini Map Aide is a mobile phone website application for visualizing atomic models and electron density maps. It emulates a small number of the functions of FRODO. It is coded in Lua, simply because that was the language that was stored in my head at the time of deciding to do this. Mini Map Aide uses a JavaScript virtual machine Fengari so that it runs as a web-browser script and can use various javascript libraries such as math.js and, most importantly, the 3D graphics library three.js. Quoting from Wikipedia (the free encyclopedia):
"Three.js is a cross-browser JavaScript library and application programming interface (API) used to create and display animated 3D computer graphics in a web browser using WebGL."
Contents |
OpenGL
WebGL uses OpenGL (Open Graphics Library) which was originally developed by Silicon Graphics in 1991 (see Wikipedia again), when a few of the last remaining PS2s were still in use, although many had been superseded by the higher powered PS300 and PS390 from E&S. I always wanted to dabble in OpenGL, but, as a non-programmer, I was put off several times by the hundreds of lines of code required to do relatively small 3D graphics tasks. Enter three.js which, thankfully, simplifies the task to the point where my interest was finally piqued into some action.
Purpose
Mini Map Aide is meant to be a completely minimal electron density map viewing aid that works on a residue-by-residue basis and could even be used by experimentalists for checking a structure in the final refinement rounds if the user has no computer access for some reason. To this end the bad regions could be recorded, e.g. by taking screen-shots, for later attention with a dedicated PC-based molecular graphics program, such as Coot. Despite its limitations, the aim is for mini map aide to be useful in research or as a teaching aid! All of the display calculations are done by a client-side script running in the user's web browser and neither the structure nor the map need to be uploaded to the server. They must, of course, be transferred to the phone beforehand by one or other means!
CCP4 Map Format
The map must be in CCP4 format and must either cover the protein molecule or fill an entire unit cell. The program should work with most of the maps which are held online in the Protein Data Bank. The steps of calculating an electron density map on a computer with the CCP4 suite installed are outlined in the online help provided at the mini map aide website. If the reflection data are available in MTZ format, these may be uploaded to the mini map aide website for conversion to a map file which can then be downloaded to your device and then loaded into the program for display. I am very grateful to Marcin Wodjyr (CCP4/Global Phasing) for allowing me to use the program sf2map from the gemmi suite on the Mini Map Aide server for this purpose.
Description of Use
Once a molecule and its map have been loaded, the program automatically rolls the structure in 3D to aid viewing, but the user can switch this to a swipe-driven rotation mode with the "Scroll" menu button. The same button switches back to "Roll" mode when required. The program will automatically centre on the Cα atom of the first amino acid in the protein and the user can step from one residue to the next with the "Next residue" button. It may also help to use the "Side chain" button which centres the display on one of the outer side chain atoms. The same button allows the user to switch back to centring on the "Main chain" Cα atom. The program has a "Go to" menu box in which the user can enter a residue number and chain ID to look at a specific residue or subunit of interest. Once the residue number is entered, the "Update" button must be clicked so redraw the display. The contour level of the map can be changed (positive values are drawn in blue, negative red) and there is a "Symmetry" option to draw the symmetry-related molecules. As for the "Go to" button, these last two options also require the "Update" button to be clicked in order for their changes to take effect. The first time the "Symmetry" checkbox is ticked, there can be a delay in redrawing the picture as the algorithm for finding all of the crystal contacts is a bit slow. Once this initial search is complete, subsequent redraws should be much quicker. Most of these options should work for polynucleotides but these have not been tested as much as polypeptides. Another possible shortcoming is that Mini Map Aide currently does not issue many warnings or error messages - the main take home message being that if something doesn't work, simply try changing one of the options or reload the webpage and try again.
Mini Map Aide attempts to mimic a tiny fraction of FRODO's protein rebuilding capabilities, albeit in a rather different way to be more amenable for use with mobile devices. On the dark display window there is a "Move" button which should always be active. When this is clicked or touched, the atoms in the residue which is currently at the centre of the display can be moved by small swipes on the screen. The moveable atoms are highlighted in a pale purple colour and a particular atom can be moved by touching it and gently swiping in one direction. The purple blob for this atom will swell for an instant to indicate that is has been selected. When the atoms have been moved to the desired locations, the geometry can be regularised and the bonds redrawn by clicking the "Tidy" button. The changes to the structure can then either be undone or saved as a new PDB file with the "Undo" and "Save" buttons, respectively.
Tidy Geometry
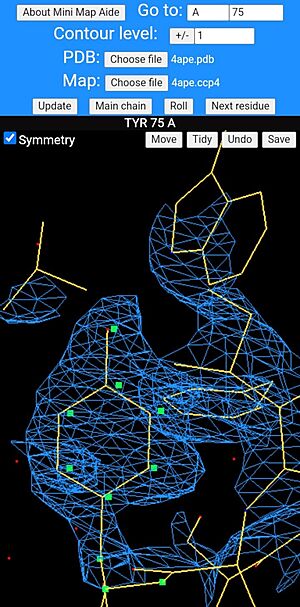
A tyrosine side chain being rebuilt in mini map aide. The moveable atoms are drawn as green squares and have been dragged slightly to the right of the ring's current position. Use of the "Tidy" button will allow the geometry to be regularised.
The option to tidy the geometry does so in a very simple stochastic way which is based on a table of approximate interatomic distances found in high resolution crystal structures of amino acids and polypeptides. These are all rounded to the nearest 0.1 Å. The method calculates all of the deviations from the ideal distances within the rebuilt residue and the atom-pair with the worst distance deviation is targeted for correction. Consider two atoms that should be covalently bonded. Since we know the direction of the bond in 3D and how much the bond length needs to increased or decreased to correct it, we could move both atoms inwards or outwards along the bond vector by half the distance violation and get a perfect bond length, at the risk of worsening the geometry of other bonds which these atoms are involved in. Instead, the algorithm takes a slightly different approach which is to take one of the two atoms at random and move it by half of the calculated distance correction in the right direction and leave the other atom unmoved. At this point the deviations from ideal geometry for the whole residue are recalculated and the worst distance violation found and corrected in exactly the same way as before. The whole process is repeated until the worst distance violation is less than 0.1 Å. The reason for choosing which of the two atoms to move completely at random is simply to stop the process becoming stuck trying to correct correlated distance violations cycle after cycle ad infinitum. It helps to prevent the algorithm getting stuck making the same two conflicting distance corrections over and over again.
Similar distance targets can be used to maintain the planarity of groups with a central trigonal atom (*e.g.* carboxylates and amides) by calculating the distance between the centroid of the three outer atoms and the central atom. This should be less than 0.1 Å and bad violations can be corrected exactly as for the interatomic distance violations, although in this case it is only the central atom (not the centroid!) which can be moved. For tetrahedral groups, such as the Cα atom of all residues except Gly and the Cβ atoms of Thr and Ile, the central atom has to be almost exactly 0.5 Å from the centroid of the three outer non-hydrogen atoms bonded to it. The vector between the central atom and the centroid is the direction along which any shifts to the central atom can be applied in 3D.
It is also possible to construct similar target positions for the atoms to help maintain the planarity and chirality of amino acids. For example in Phe and Tyr, the Cβ - Cγ bond will be parallel, well within experimental error, to the Cδ - Cε bonds in the aromatic ring. Given the slight difference in length between single and double bonds (which is known from atomic resolution studies), we can calculate a target position for each of the Cε atoms from the direction of the Cβ - Cγ bond and the current Cδ atom positions. The distance between each Cε atom and its target position is fed into the list of distance violations and will be corrected by the algorithm when it becomes the worst one. Longer-range constraints, such as ensuring the phenolic C-OH bond of Tyr is also parallel to these bonds helps to maintain planarity. In addition, we can correct main chain and side chain chirality errors by using math.js to calculate the signed volume of these groups. When the chiral volume is negative, the group needs to be inverted. The program uses a fairly crude way of doing this which is to move the central atom to the centroid of the three outer non-hydrogen atoms and one of the outer atoms is shifted the same distance but in exactly the opposite direction in 3D. Subsequent iterations to correct the interatomic distances seem to sort out any remaining anomalies introduced by flipping the hand of the group in this way.
Whilst this is a far cry from molecular mechanics (something best left to computational chemists!) it does seem to give reasonable geometry to the amino acids and usually takes only 1 or 2 seconds at most on a humble Android smartphone.
In future versions, it is planned to allow simple mutations to be made.
See Also
- Molecular visualization in the 1980s where FRODO is described.
- History of Macromolecular Visualization
- X-ray crystallography
- Electron density maps
Content Attribution
This article was written by Jonathan Cooper, Professor Emeritus at University College, London. (Eric Martz simply put it into Proteopedia, but had no authorship role. Angel Herráez styled the images with captions.)