Missing residues and incomplete sidechains
From Proteopedia
Most empirical models are missing standard amino acid or nucleotide residues that were present in the experimental material, and/or sidechain atoms, and therefore are often missing charges present in the intact molecule. These missing parts make errors in the shape of the molecule as well as the distribution of charges, hydrophobic and polar regions, and often there are missing salt bridges and cation-pi interactions. FirstGlance in Jmol makes it less likely, compared to other popular viewers, to overlook missing parts. AlphaFold models, which lack no atoms (other than hydrogen), should be analyzed alongside empirical models in order to see complete structures.
Missing Residues
In about 80%[1] of the Empirical models in the PDB, some residues (amino acids or nucleotides) that were present in the experimental material are absent (have no coordinates) in the empirical model. The breakdown by method as of February, 2025:
- 83% of X-ray crystallography entries have missing residues.
- 14% of solution NMR entries have missing residues.
- 93% of electron microscopy entries have missing residues.
X-ray crystallography gives a clear electron density map only where every molecule in the protein crystal has the same conformation. Usually, some parts of the molecule vary in conformation between copies in the crystal, that is, some regions are disordered. The same may occur with protein molecules on a cryo-electron microscopy grid. These disordered portions of the molecule are not clearly resolved in the density map used to construct the structure model. Without density to guide where to place these residues, the experimenter omits them from the model. These are called missing residues. It is very common for a few residues at the ends of protein chains to be missing in the atomic model. (Example: 5 residues are missing from the carboxy terminus of the protein in 1ijw.) In addition to disorder rendering residues "invisible", residues can actually be lost by proteolysis during purification, or radiation damage during X-ray crystallography can sometimes remove atoms[2], and conformations can be affected by crystallization conditions[3].
To emphasize, in most cases, the missing residues were believed to be present in the experimental material, but are absent in the resulting atomic model. Missing residues are distinct from intentionally deleted residues that were not present in the experimental material due to engineering of the cloned and expressed protein.
Get The Best Model Available
Before you spend time working with an empirical model, make sure you have chosen the highest resolution and most complete model available. Search for sequence-identical (or nearly identical) models. For a step by step guide, see How to find a protein's best structure. Things will be much easier if the sequence numbers in the empirical model are the same as the UniProt numbering. If not, consider renumbering the PDB file.
Missing Ends of Chains
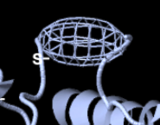
Unlike other viewers, FirstGlance in Jmol ensures that you are aware of missing ends by marking them with spherical "empty baskets". In the example shown below, 2ace, the 3 missing N-terminal residues DDH have net negative charge.
|
2ace amino-terminus missing 3 amino acids |
P04058 AlphaFold: | ||||
|---|---|---|---|---|---|
|
FirstGlance "empty basket". | |||||
Missing Loops
FirstGlance ensures that you are aware of missing loops with an ellipsoidal "empty basket". Other viewers use a dotted line, which is easier to overlook when viewing the entire structure. Empty baskets are easily hidden[4].
|
2ace missing 5 amino acid loop |
P04058 AlphaFold: | ||||
|---|---|---|---|---|---|
|
FirstGlance "empty basket" (see #below)[5] | |||||
See For Yourself
- 2ace in FirstGlance in Jmol
- 2ace in Mol*
- 2ace in iCn3D
- (PyMOL and ChimeraX require that you download stand-alone applications.)
AlphaFold models have no missing atoms
When all empirical models of the protein of interest have missing atoms, the best way to get a model without missing atoms is to download the AlphaFold model. The downloaded PDB file can then be uploaded to FirstGlance. iCn3D will accept the UniProt sequence ID to retrieve the AlphaFold model directly.
Models from the AlphaFold Database are limited to single-chain structures without ligands, but starting in 2024, see How to predict structures with AlphaFold for servers that can predict multiple-chain structures (protein, DNA, RNA) with ligands or modified residues.
The AlphaFold model will usually be nearly identical to the empirical model (you should verify this by superposition), but will include the missing residues/atoms. Structures of missing loops will be predicted by AlphaFold, but typically with lower confidence (lower pLDDT) than the remainder of the structure. Many long loops that are missing in empirical models are actually intrinsically disordered, in which case those loops predicted by AlphaFold will have very low confidence and be meaningless.
Why Missing Residues Matter
Where residues are completely missing in empirical models, the shape of the molecule will be incorrect, and when charged residues are missing, the distribution of charges will be incorrect, as well as the distribution of hydrophobic and polar patches. Salt bridges and cation-pi interactions may be missing.
Example: 5nyp
5nyp is a bacterial protein believed to be an ancestor of the 20S proteasome[6]. All sequence numbers given below are UniProt numbering. Subtract one to match numbering in 5nyp.
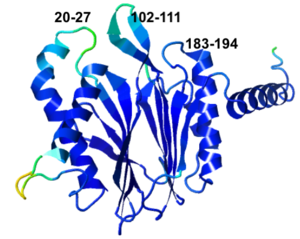
- 5nyp has 3 missing loops, clustered on one face. Their lengths are 8, 10, and 12 residues (20-27, 102-111, 183-194).
- None of the 3 missing loops are predicted to be intrinsically disordered by RCSB[7]. The first missing loop is predicted to be disordered by flDPnn2[8]. The other two are not. But flDPnn2a predicts disorder for 71-83, which is not missing.
- The missing loops include 7 charged amino acids (4–, 3+).
- Although the authors did not detect proteolytic activity with the substrates tested, the putative catalytic triad residues Thr2, Asp18, and Lys33 (UniProt numbers) are present.
- When present in the AlphaFold model, two of the missing loops partially obscure the putative catalytic triad.
- Two salt bridges are missing in 5nyp that are present in the AlphaFold model. UniProt Glu107 (present) forms a salt bridge with missing Arg99. UniProt Asp183 forms a salt bridge with arg187 -- both are missing in 5nyp.
|
Three Missing Loops in 5nyp. Snapshots from FirstGlance. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
5nyp: missing loops (in front) as ellipsoidal "empty baskets"[9].
|
5nyp: yellow halos on putative catalytic triad, showing proximity to missing loops (in front). |
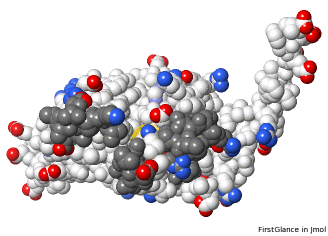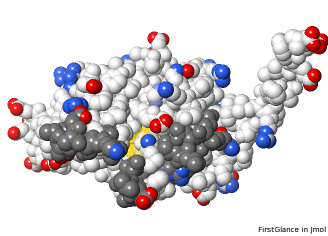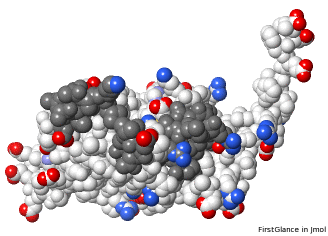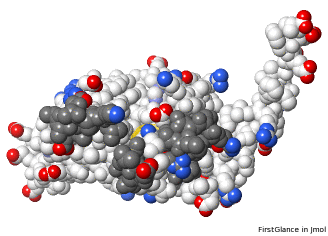
AlphaFold-predicted structure for F8JB59. Loops missing in 5nyp are at top.
Confidence (>70 is "confident"): | |||||||||
AlphaFold Prediction Superposed on 5nyp
|
The three missing loops are present in the AlphaFold prediction for UniProt F8JB59. FATCAT superposed the AlphaFold prediction onto all 213 alpha carbons of 5nyp with RMSD 2.4 Å. The FATCAT-generated morph between the two models shows their similarity. Their similarity shows that the AlphaFold model is largely correct. |
AlphaFold Prediction Superposed on 5nyp |
Morph Between Models |
|---|---|---|
Missing loops at top. |
Missing Loops Obstruct Catalytic Site
|
Missing loops affect the shape of the molecule. The putative catalytic site is exposed on 5nyp, but when the 3 loops are added in AlphaFold's prediction for UniProt F8JB59, they partially obstruct the putative catalytic site. |
AlphaFold-Predicted Structure | |
|---|---|---|
Charged atoms colored by FirstGlance[10]: Positive +, Negative –. Putative catalytic triad: yellow halos. Loops predicted by AlphaFold in dark gray. | ||
Missing Charges in 5nyp
|
The three missing loops include 4 negatively-charged amino acids, and 3 positively charged ones. Their absence affects the distribution of charge on the surface, as shown in these electrostatic potential maps generated by iCn3D (from a link in the Views tab in FirstGlance). |
5nyp: 3 loops with 7 charges are missing. |
AlphaFold-Predicted Structure for F8JB59: No missing charges. |
|---|---|---|
Electrostatic potential maps by iCn3D (from a link in the Views tab of FirstGlance):
| ||
Missing Salt Bridges in 5nyp
|
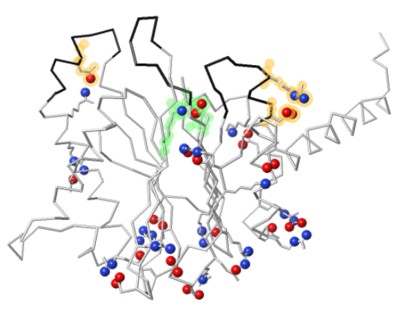
Two salt bridges are missing in 5nyp as a result of two of the missing loops. At left: Missing UniProt Glu107 forms a salt bridge with Arg99 (present). At right: UniProt Asp183 forms a salt bridge with Arg187 -- both are missing in 5nyp. A salt bridge also forms between two of the putative catalytic triad residues, Lys31 and Asp169. (All 3 triad residues are present in 5nyp.) |
AlphaFold-Predicted Structure for F8JB59. |
|---|---|
Salt bridges rendered by FirstGlance[11]: Positive +, Negative –. Yellow halos: Charges missing in 5nyp. Green halos: Putative catalytic triad, present in 5nyp. Black backbone: loops missing in 5nyp. |
Incomplete Sidechains
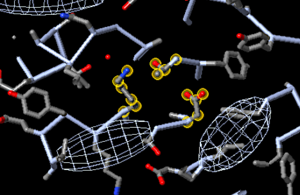
In about half[1] of the Empirical models in the PDB, some residues have coordinates missing for some of their sidechain atoms, due to local disorder. But their main chain atoms are present in the model; thus, they are residues with incomplete sidechains. For example, the long sidechain of a lysine on the surface of a protein may have too blurry an electron density to indicate its position. In some cases, the model builder may give that sidechain coordinates with high temperatures, or low or zero occupancy (example: Arg321 in 2ade[12]). In other cases, the model builder simply omits the coordinates for the sidechain, so the aforementioned surface lysine may have the sidechain of an alanine (example: Lys498 in 2ace).
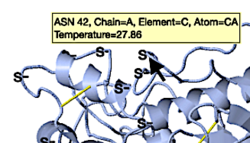
Among the five viewers shown below, only FirstGlance alerts you to incomplete sidechains in its initial view. It marks them S—. FirstGlance and iCn3D are the only ones that show disulfide bonds in their initial views, although, as you can see in the snapshots below, FirstGlance makes them more obvious. The S— labels are easily hidden when desired[13].
|
Initial views of incomplete sidechains in 2ace | ||||
|---|---|---|---|---|
|
FirstGlance marks incomplete sidechains with S—, and shows disulfide bonds. |
iCn3D shows disulfide bonds. | |||
Do incomplete sidechains matter?
Obviously, incomplete sidechains matter when they occur near a region of interest in an empirical model. The easiest way to get a model without incomplete sidechains is to use the AlphaFold model, bearing in mind that residue positions and sidechain rotamer orientations may be inexact.
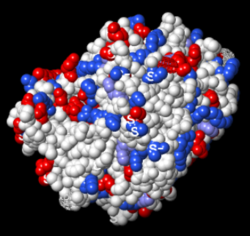
Atoms colored by charge
When the charged atoms are missing in incomplete sidechains, there is little effect when spacefilling atoms are colored by charge in FirstGlance or iCn3D, because they color the entire residue by charge. FirstGlance initially colors only charged atoms. Where the charged sidechain atoms are missing, FirstGlance colors the remainder of the residue in a lighter color, with a checkbox option to color all atoms in charged residues, as does iCn3D.
|
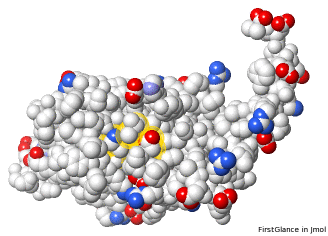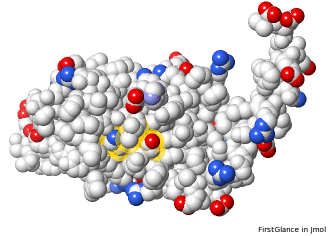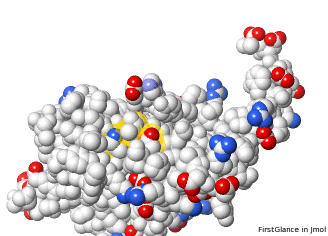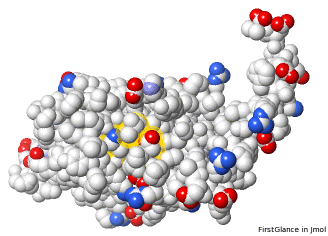
Atoms Colored By Charge: 2ace With Incomplete Sidechains, Positive +, Negative – | |||
|---|---|---|---|
|
FirstGlance: orientation for all snapshots. |
FirstGlance default[10]: charged atoms, light colors for incomplete sidechains. |
FirstGlance charged residues checked. | |
(No "single button" to color spacefilling atoms by charge was evident in Molstar or ChimeraX. In ChimeraX, the commands to do this can be saved as a "preset".)
Surface colored by electrostatic potential
When either ChimeraX or iCn3D color a surface by electrostatic potential, incomplete sidechains affect the map. Therefore, it is critical to use the AlphaFold-predicted structure that has neither missing residues nor incomplete sidechains.
|
Surface Colored By Electrostatic Potential vs. Incomplete Sidechains for 2ace, Positive +, Negative – | |||
|---|---|---|---|
|
Structure Visualized |
2ace with incomplete sidechains. |
AlphaFold-predicted structure for P04058, with both ends trimmed.[18][19]. | |
Orientations for all snapshots are the same as in the previous table. | |||
How to avoid overlooking missing residues or incomplete sidechains
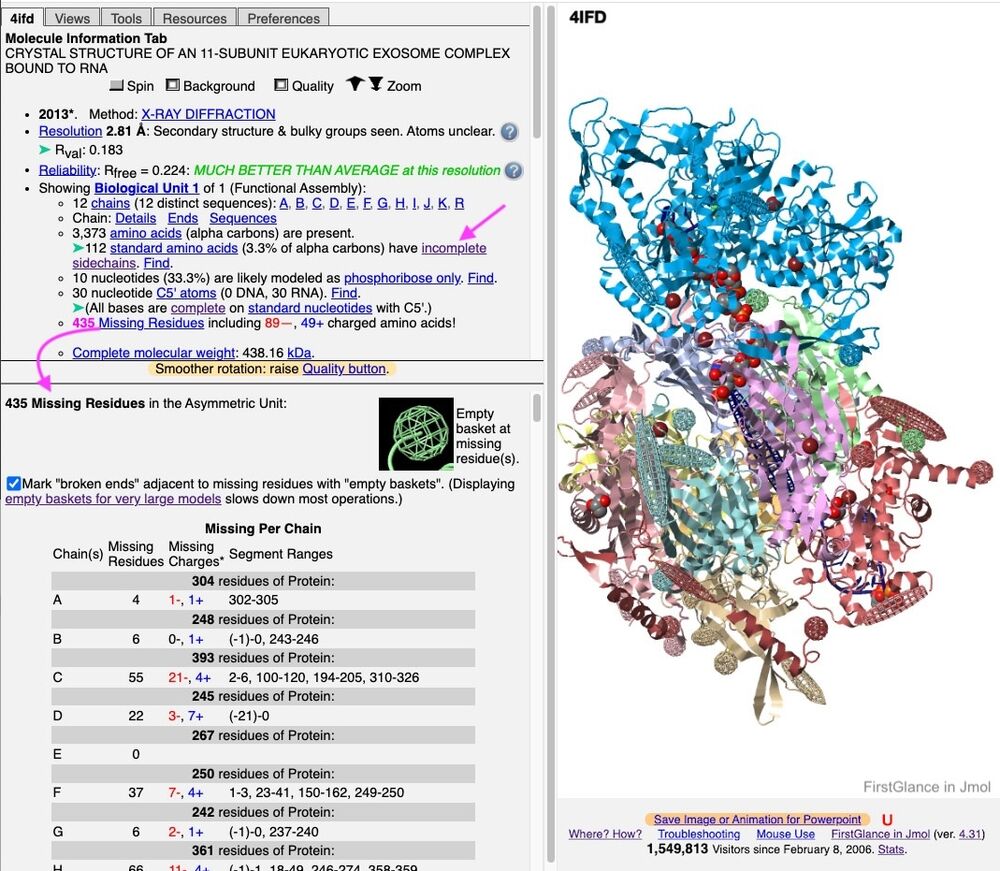
Regions with missing residues are clearly marked with "empty baskets" when a PDB ID model is displayed in FirstGlance in Jmol, as shown above. FirstGlance reports the total number missing, and the resulting number of missing charges. And it offers a detailed report (see snapshot below), which also lists all the missing residues (from PDB file REMARK 465).
FirstGlance also marks incomplete sidechains with S–, and (when you opt to Show More Details) provides a count of Incomplete Sidechains (see snapshot below). Clicking Find/List at that count enables you to list all the residues with incomplete sidechains.
Example in FirstGlance 4ifd: (When there are >100 incomplete sidechains, the S– labels are initially suppressed to reduce clutter.)
Missing residues and atoms lists in PDB and mmCIF files
PDB Format
Quoting from the legacy PDB Format Description:
"REMARK 465 lists the residues that are present in the SEQRES records but are completely absent from the coordinates section."
SEQRES lists only protein and nucleic acid linear polymer residues, not carbohydrate or other residues of hetero atoms. Therefore, missing residues of polysaccharides or other HETERO compounds are not listed in the PDB file.
Similarly,
"REMARK 470: Non-hydrogen atoms of standard residues which are missing from the coordinates are listed. Missing HETATMs (atoms within hetetrogen groups) are not listed here."
Example:
- Look for REMARK 465 and REMARK 470 in the PDB file for 2ACE.
- Look for empty baskets and S— labels for 2ACE in FirstGlance.
Zero occupancy vs. missing atoms
Atoms with zero occupancy have coordinates, and therefore are not listed as missing. For example, 1nsa (1997, resolution 2.3 Å) has 64 distal sidechain atoms with zero occupancy on the surface of the protein. 3dxx (2009, resolution 2.1 Å) has 27 distal surface sidechain atoms with zero occupancy. Neither has REMARK 470. Additional zero occupancy cases are 8pch and 3b1d. Alternatively, these atoms could have been left missing (giving incomplete sidechains), a more typical treatment of weak electron densities.
FirstGlance in Jmol tabulates a summary of occupancies, and can list partial occupancies in spreadsheet-ready form, for example
- 1NSA in FirstGlance: Click Show More Details, then scroll down in the upper left panel to click Occupancy.
mmCIF Format
PDB to PDBx/mmCIF Data Item Correspondences provides no information about REMARK 465 or REMARK 470.
Although pdbx_missing_residue_list and _pdbx_missing_atom_poly are in the mmCIF dictionary, they are described as not used in current PDB entries.
It appears that these lists are in pdbx_unobs_or_zero_occ_residues and pdbx_unobs_or_zero_occ_atoms. However, these mmCIF categories lump together missing (unobserved) and zero occupancy cases, unlike the separation of these two categories in the PDB format.
Examination of mmCIF files suggests that missing residues are also indicated by "?" in certain data items of category _pdbx_poly_seq_scheme.
Incomplete Sidechain Repair
If you need to add the missing atoms for incomplete sidechains without changing the positions (coordinates) for any existing atoms, here are two methods:
- Use the free Protein Repair & Analysis Server.
- Simply load the PDB file into free Swiss PDB Viewer, then save it. This program automatically adds missing sidechain atoms. However, the program will not run on Mac OS 10.15 or higher. It will not run on Silicon CPU Macs.
See Also
Notes & References
- ↑ 1.0 1.1 Percentages are based on searches for REMARK 465 and REMARK 470 at OCA, and searches for unobserved residues and unobserved atoms at RCSB.
- ↑ Weik M, Ravelli RB, Kryger G, McSweeney S, Raves ML, Harel M, Gros P, Silman I, Kroon J, Sussman JL. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000 Jan 18;97(2):623-8. PMID:10639129
- ↑ Dym O, Song W, Felder C, Roth E, Shnyrov V, Ashani Y, Xu Y, Joosten RP, Weiner L, Sussman JL, Silman I. The Impact of Crystallization Conditions on Structure-Based Drug Design: A Case Study on the Methylene Blue/Acetylcholinesterase Complex. Protein Sci. 2016 Mar 14. doi: 10.1002/pro.2923. PMID:26990888 doi:http://dx.doi.org/10.1002/pro.2923
- ↑ See Hiding empty baskets.
- ↑ The "S-" indicates that Lys491 has an incomplete sidechain, extending only to the gamma carbon.
- ↑ Vielberg MT, Bauer VC, Groll M. On the Trails of the Proteasome Fold: Structural and Functional Analysis of the Ancestral beta-Subunit Protein Anbu. J Mol Biol. 2018 Feb 2. pii: S0022-2836(18)30007-X. doi:, 10.1016/j.jmb.2018.01.004. PMID:29355501 doi:http://dx.doi.org/10.1016/j.jmb.2018.01.004
- ↑ Erdős G, Dosztányi Z. Analyzing Protein Disorder with IUPred2A. Curr Protoc Bioinformatics. 2020 Jun;70(1):e99. PMID:32237272 doi:10.1002/cpbi.99
- ↑ Wang K, Hu G, Basu S, Kurgan L. flDPnn2: Accurate and Fast Predictor of Intrinsic Disorder in Proteins. J Mol Biol. 2024 Sep 1;436(17):168605. PMID:39237195 doi:10.1016/j.jmb.2024.168605
- ↑ There is a spherical blue "empty basket" on the amino terminus because Met1 is missing. This spherical empty basket was removed in the image showing the putative catalytic triad.
- ↑ 10.0 10.1 To color a protein by charge in FirstGlance, click Charge in the Views tab.
- ↑ To display and list all salt bridges in a protein, click Salt Bridges in the Tools tab.
- ↑ In 2ade, the temperature of the distal nitrogens in Arg321 is 79, while the average temperature is 31.
- ↑ See Labels on Atoms: S—, X, D, ?.
- ↑ To color a protein by charge in iCn3D: Style, Protein, Spheres; then Color, Charge.
- ↑ To generate an electrostatic potential map in ChimeraX, with nothing selected, click electrostatic in the Molecule Display tab. To intensify the colors, reduce the default range (-10 to 10), for example to -5 to 5, select the desired protein, then enter this command: coulombic sel range -5,5.
- ↑ Electrostatic potential maps were created in iCn3D using its Analysis, Delphi Potential, Surface with potential, with default settings except the max potential was set to 6 kT/e.
- ↑ To fill in incomplete sidechains in 2ace, the PDB file was loaded into Swiss-PDBViewer, then saved. It completes sidechains automatically. It does NOT fill in missing residues.
- ↑ The ends absent from the protein crystallized for 2ace were deleted using a Plain text editor. The signal sequence 1-21 was deleted from the N terminus. The C terminus starting with 559 was deleted; it is a tetramerization domain (see next reference).
- ↑ Dvir H, Harel M, Bon S, Liu WQ, Vidal M, Garbay C, Sussman JL, Massoulie J, Silman I. The synaptic acetylcholinesterase tetramer assembles around a polyproline II helix. EMBO J. 2004 Nov 10;23(22):4394-405. Epub 2004 Nov 4. PMID:15526038 doi:7600425