User:Eric Martz/Introduction to Structural Bioinformatics 2016
From Proteopedia
How to visualize, understand and share/present 3D protein molecular structures
by Eric Martz, 2016
for
Microbiology 497L: Advanced Microbiology Lab Techniques
University of Massachusetts, Amherst MA USA
Get here via Moodle or with 497L.MolviZ.Org
I. Getting Started in the FAC444
- Use FIREFOX or Safari.
(DO NOT USE Chrome because molecule rotation will be slow/jerky. Internet Explorer and Edge are even worse with this software.)
- Go to our syllabus: 497L.MolviZ.Org.
- Now you can see this document in your browser. Go to Atlas.MolviZ.Org.
- In the Atlas, choose any molecule deemed Straightforward and click on the link to FirstGlance. After a minute or so to load, you should see a rotating molecule.
- If you have any difficulty or the molecule does not appear, ask for help!
II. Goals
1. Review principles of protein 3D structure.
2. Choose an existing experimentally-determined 3D protein structure model to investigate.
3. Learn how FirstGlance in Jmol makes it easy to see structure-function relationships in the protein you chose.
4. Write a report including snapshots of your protein that illustrate your answers to the questions below. (Your report will be in the form of Google Slides emailed to emartz@microbio.umass.edu. You will not present your report in class.)
III. Protein Structure and Structural Bioinformatics
|
|
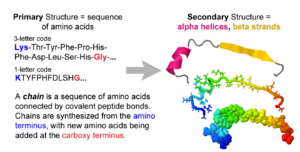
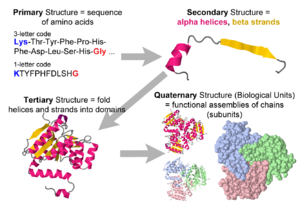
- 1. Amino acid sequence + protein chain conformation = protein function.
- A. Why do we care about protein 3D structure?
- B. Conformation can be a stable fold or intrinsically unstructured. Both commonly exist in the same protein molecule.
- C. Conformation is specified by sequence.
- Folded domains fold spontaneously (Anfinson, 1960's[1]), or with the help of chaperonins.
- The denaturation (unfolding) of a folded protein domain destroys its function.
- 2. Backbone Representation.
- A. Backbone Representations
- B. Small Protein in FirstGlance (use the Views Tab: Vines, Cartoon)
- 3. Structure Knowledge.
- A. Although sequence specifies fold, scientists cannot yet predict the fold from the sequence. Therefore, fold must be determined by empirical (experimental) methods. The most common methods for determining the 3D structure of a protein molecule are:
- X-ray crystallography, 88%.
- Result is a single model representing the average of the molecules in the crystal.
- Resolution reflects the degree of order or disorder in the crystal.
- X-ray crystallography gives no models for intrinsically unstructured loops or molecules.
- Nuclear magnetic resonance (NMR) in aqueous solution, 9%.
- NMR is limited to small proteins (30 kD or smaller; median NMR in PDB is 10K; median X-ray is 50K).
- Result is an ensemble of models consistent with the data. Examples: 2bbn
- High resolution cryo-electron microscopy, 0.8%.
- A. Although sequence specifies fold, scientists cannot yet predict the fold from the sequence. Therefore, fold must be determined by empirical (experimental) methods. The most common methods for determining the 3D structure of a protein molecule are:
- B. These methods are difficult and expensive. Less than 10% of proteins have known structure.
- C. All published, empirically determined 3D macromolecular structure models are available from the Protein Data Bank (PDB; pdb.org; About the PDB).
- E. Crystallographers publish the asymmetric unit of the crystal. It may be identical with the biological unit (the functional form of the molecule), or it may be only part of the biological unit, or it may contain multiple copies of the biological unit. See examples.
- Interchain contacts that occur in the asymmetric unit, which are absent in the biological unit, are an artifact of crystallization, termed crystal contacts.
- E. Crystallographers publish the asymmetric unit of the crystal. It may be identical with the biological unit (the functional form of the molecule), or it may be only part of the biological unit, or it may contain multiple copies of the biological unit. See examples.
IV. Choose a Molecule to Explore
- Choose a molecule to use for your report.
- Each student should choose a different molecule.
- Be sure to note the 4-character PDB code of the molecule you choose. The PDB code makes it easy to retrieve the molecule and information about it.
- Report the PDB code you chose to the instructor to make sure it is not already taken.
- It must have protein.
- It will be more interesting if it contains some non-protein: ligand, or DNA or RNA.
- X-ray results should have resolution of 3 Å or better.
- Here are some ways to find a protein with known structure:
- Recommended: Atlas of Macromolecules (Atlas.MolviZ.Org). Choose a "Straightforward" or "Challenging" (not "Enormous") molecule that has protein and ligand.
- Molecule of the Month at the PDB. Look for PDB codes in the article, and use FirstGlance to view them.
- Topic Pages in Proteopedia, or its Table of Contents.
- Random PDB Entry in Proteopedia (see Random at top left of this page in the navigation box).
- Search by molecule name or amino acid sequence at www.pdb.org, but remember that less than 10% of proteins have known structure. See also Practical Guide to Homology Modeling which includes instructions for finding empirical 3D models for a protein sequence.
V. Explore Your Molecule
FirstGlance in Jmol
The main tool we will use is FirstGlance in Jmol: FirstGlance.Jmol.Org. (To google it later, use the single word (no space) firstglance.)
- Use Firefox (or Safari). The molecule will rotate slowly with jerky jumps in other browsers. Internet Explorer and Edge are especially bad for Jmol.
- Enter your 4-character PDB code at FirstGlance, and you should see the molecule you have chosen.
- Get familiar with what the molecule information tab tells you. Ask about anything you don't understand.
- In the Views tab, there are 10 links at the top that show you different aspects of the molecule. Try them all, as well as any of the other tools in FirstGlance that interest you.
-
FirstGlance does NOT use Java unless you tell it to. Using Java will make larger proteins load faster, rotate more smoothly, and change views quicker. Use Firefox or Internet Explorer (or Safari) for Java. Chrome and Edge do not support Java. In your Java-compatible browser, display a molecule in FirstGlance, and then click on the Preferences tab in FirstGlance. See Installing and enabling Java.
Here are two Views in FirstGlance that will be used in your report:
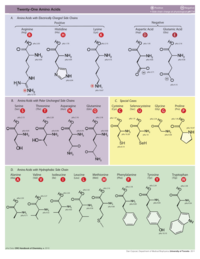
A. Hydrophobic/Polar
- Water-soluble proteins have polar/charged amino acids nearly everywhere on their surfaces (Examples: small 2hhd, large 1igy). Patches of hydrophobic amino acids on the surfaces of soluble proteins are usually less than ~10 å in their smaller diameter, and usually recessed.
- Hydrophobic surface patches may be buried in chain-to-chain contacts -- check the biological unit (example: lac repressor homodimer).
- Large, protruding hydrophobic surface areas (>25 Å in their smaller diameter) may indicate transmembrane proteins (insoluble). Examples:
- 7ahl
- showing bilayer boundaries (click on "Jmol"; ligand toggles boundaries).
- 1bl8
- Gramicidin Channel in Lipid Bilayer.
B. Charge
Most proteins have roughly equal numbers of positive and negative charges intermixed on their surfaces. Surface patches of exclusively positive charge often bind nucleic acids (negatively charged because of their phosphates). For example, examine the protein surface charges where the gal4 transcriptional regulator binds DNA (1d66).
VI. Report Slides
Answer the questions below in slides, using Google Slides.
- Name your report YourLastName-497L, for example Sandler-497L. If the name of your report does not begin with your family name, you will lose 2 points. When completed, email a link to your report to emartz@microbio.umass.edu for grading.
- (While viewing your report slides, click the blue Share button at the upper right, then Get shareable link, and paste the link into the email.)
- You will not be asked to present your report in class.
- Each slide MUST be labeled at the top with its section number, e.g. Section 1.
- Each Section below may be answered in a single slide, or multiple slides. For example, suppose you want to show two snapshots for Section 3, and make separate comments. You may choose to use two slides, labeled Section 3A and Section 3B.
- Additional work beyond the minimum required may earn extra credit.
- Due date: midnight Thursday March 3.
This is not a test. It is to help you learn by doing. Ask for help!
Example of a Completed Report (You may import these slides into a new presentation of your own, and use them as a templates, putting in your own content.)
Section 1: Identity
- The label Section 1 at the top (and so forth for every slide).
- Your name.
- Your major; grad students, give the name of your grad program (Micro, MCB, etc.) and whose lab you work in.
- Your PDB identification code.
- The name of your molecule.
- The function of your molecule (briefly, one sentence).
- The experimental method and resolution (or number of models for NMR). Available in the molecule information tab in FirstGlance.
- A snapshot of your molecule.
is also linked at the bottom left in FirstGlance. |
Section 2: Composition
- The number of
- Protein chains (What does "chain" mean?)
- DNA chains
- RNA chains
- Available in the molecule information tab in FirstGlance: Chain Details. You can identify a residue in any chain by touching it with the mouse (spinning off!). DNA residues are DA, DG, DC, DT while RNA residues are A, G, C and U.
- Ligands and Non-Standard Residues: Give the one to three-letter abbreviations and full names for all ligands and non-standard residues. If none, so state. (Standard residues)
- The molecule information tab in FirstGlance lists the 1 to 3-letter abbreviations for each ligand and non-standard residue, and their full names. Click on an abbreviation to locate that entity in the model. See also Composition in FirstGlance's Views tab.
Section 3: Evolutionary Conservation
See Introduction to Evolutionary Conservation. (Example 4d7b: ConSurf, Result)
Does your molecule have a highly conserved region? If so, what is its function? If there is no highly conserved region, is there a highly variable region? Show two snapshots illustrating a highly conserved region, and a contrasting region.
See How to see conserved regions.
- If Proteopedia lacks a pre-calculated Evolutionary Conservation for your molecule, and you do your own calculation at the ConSurf Server, be sure to include the address of the ConSurf result in your report slide!.
Section 4: Hydrophobic/Polar
Do you think your molecule is water soluble? Support your conclusion with a snapshot. Be sure to use the Hydrophobic/Polar view from FirstGlance in a snapshot. Optionally, you may show other views in other snapshots.
Section 5: Hydrophobic Core
Are there hydrophobic cores in your molecule? For soluble proteins, expect a hydrophobic core in each domain. Support your conclusion with a snapshot. Be sure to use the Hydrophobic/Polar view from FirstGlance and turn on the Slab button.
Section 6: Charge
Are there any areas on the surface of your molecule with only positive (or negative) charges? Show snapshots illustrating your conclusions. Be sure to use the Charge view from FirstGlance in your snapshots.
Section 7: Cation-Pi Interactions
Show a snapshot of an energetically significant cation-pi interaction. Include a distance monitor in your snapshot. Also paste in the report from CaPTURE confirming its energetic significance. The cation-pi interaction tool, and instructions for measuring distances, are in the Tools Tab.
Section 8: Biological Unit
In FirstGlance, in the molecule information tab click Biological Unit. (It is also in the Resources Tab.)
How many total polymer chains (protein + DNA + RNA) are in the asymmetric unit? The Biological unit?
Show side-by-side two snapshots comparing the asymmetric unit with the biological unit. The Cartoon representation in FirstGlance is best for these snapshots. Make sure to label which is which.
Section 9: Animation from Polyview-3D
Minimal steps to make an animation:
- Go to Polyview-3D.
- Enter your PDB code in the PDB ID slot near the top. (If the slot is not visible, open the section Source of Structural Data.)
- Change "Type of request" from "Single slide" to "Animation". It is under the Image Settings section near the bottom.
- Click any "Preview" link.
- Optional: If you want to modify the orientation or zoom of the molecule, click on View by Jmol / Set orientation under Quick links at the upper left of the page. Use the mouse to rotate and zoom in Jmol. Then click the Set and close button.
- Optional: If you want to change the colors, hide portions of the molecule, emphasize certain residues, etc. feel free to try out these options in the form, using Preview to check your results.
- In the "Animation Settings" section at the bottom of the page, set Delay to 10/100.
- Change "Angle step" to 5 degrees.
- Check "Rocking".
- Change "angle range" for rocking to 30 degrees.
- Click "Submit".
The above steps are the minimum for an animation that avoids putting a heavy load on the server. Feel free to try other options, but while the class is in session, please don't make a large (>300 pixel) animation, or increase the angle range, or decrease the angle step size. Otherwise, the server may get overloaded and take a very long time to produce results. Optional: After class is over, feel free to submit more demanding jobs. If you highlight specific residues, please explain why.
After your animation is completed and appears in the PolyView-3D web page:
- Right-click (Mac: control-click, or trackpad 2-finger click) on the animation in the Polyview-3D web page, and select Save Image As ...
- Save the image to the Desktop.
- Drag the image file (filename ending in .gif) from the Desktop and drop it into a slide.
- In Google Slides, the animation should move immediately.
Examples of Slides with Polyview-3D Animations (These slides are only to show you what is possible. These are not in your assignment.)
Section 10 - Contacts/Non-covalent Bonds
Example: 4d7b.
- Click Contacts in the Tools Tab in FirstGlance.
- Change target selection to Residues/Groups.
- Click on something small to select it as a "target", such as a ligand, or a single amino acid. Choose an amino acid with an uncharged polar side chain, such as Ser, Thr, Asn, Gln, Tyr, His.
- Click the link to Show atoms contacting target.
- Click Center contacts.
- Uncheck Backbones.
- Click the 4th thumbnail image
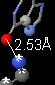 to display the contacts as balls and sticks colored by element. The element color key is at the bottom of the Contacts help panel in FirstGlance.
to display the contacts as balls and sticks colored by element. The element color key is at the bottom of the Contacts help panel in FirstGlance.
- Zoom in (and click Return to Contacts if necessary).
- Uncheck all categories of non-covalent bonds.
- Check hydrogen-bonded non-water. (Review hydrogen bonds.)
- Double click the hydrogen bond donor and acceptor atoms to insert a distance monitor.
Describe the moiety you selected as a target. Include a snapshot showing exactly one hydrogen bond. Be sure to identify the two entities (amino acids, nucleotides, ligand) by name, chain, and sequence number. I need enough detail to be able to reproduce what you are reporting.
Section 11 - How Structure Supports Function
Write a brief description in your own words (avoid plagiarism!) of how the structure of this protein supports its function. Doing some online research will strengthen your description.
Include links to supporting references. Wikipedia can be cited, but authoritative sources, such as peer-reviewed scientific journal articles or government websites, will have more weight.
Your description should be at least 75 words. More work, if well done, will earn more credit.
Section 11 in the Sample Report is longer than the minimum required, but illustrates the sort of thing that could be done if you can spend the time.
VII. See Also
- User:Eric Martz/Introduction to Structural Bioinformatics, a list of courses and workshops at various levels, including earlier versions of this segment.
VIII. Notes and References
- ↑ For a brief overview of Anfinson's protein folding experiments in the 1960's, see the first paragraph at Intrinsically Disordered Protein.