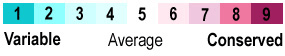ConSurfDB vs. ConSurf
From Proteopedia
|
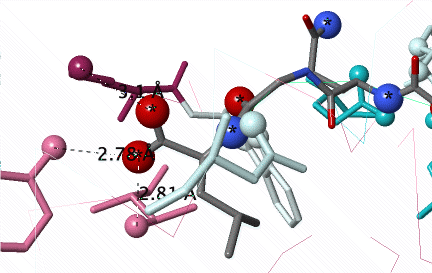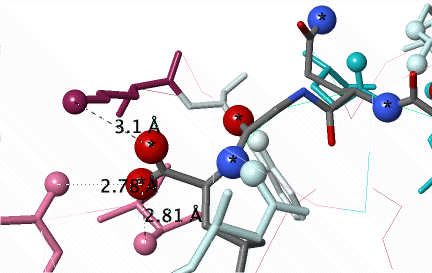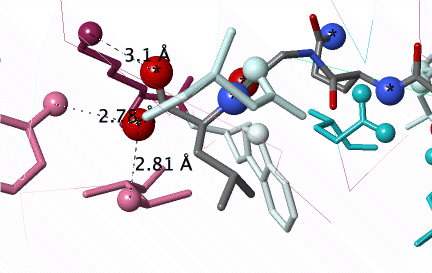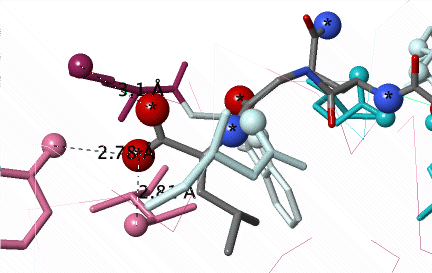
Conservation of amino acids non-covalently interacting with a peptide (C N O) in the groove of Major Histocompatibility Protein Class I (2vaa). Conservation was not revealed until an #Average Pairwise Distance of 0.31 was achieved in a customized ConSurf Server job. DETAILS BELOW. |
Evolutionary Conservation is introduced at Introduction to Evolutionary Conservation, and treated in somewhat greater depth in the article Conservation, Evolutionary. These describe how conservation patterns in 3D can help to identify functional sites in proteins. Proteopedia displays conservation patterns pre-calculated by ConSurfDB, when available. These are usually based on broad protein families that include sequences of proteins with multiple functions. Consequently, they usually obscure conservation present in a family of proteins with a single function (see Caveats).
The present article explains how to use options available at the ConSurf Server to reveal conservation within a group limited to proteins with a single function. Several examples are presented. The mechanisms utilized by ConSurfDB and ConSurf Servers are also summarized.
Contents |
The Two ConSurf Servers
There are two ConSurf servers:
- ConSurfDB:
- Since 2022, ConSurfDB results are NOT compatible with FirstGlance in Jmol. Visualizing and analyzing conservation results in FirstGlance has many advantages. Use the ConSurf Server for compatibility with FirstGlance.
- In July, 2024, ConSurfDB had not been updated with new entries in the Protein Data Bank since mid-2022. To look for more recent updates, at ConSurfDB Home, click More.
- Has pre-calculated results for every chain in the PDB.
- Proteopedia's Evolutionary Conservation resource displays results from ConSurfDB.
- Results typically obscure some conservation related to a protein's function because the analysis typically included proteins of multiple functions (see ConSurfDB Often Obscures Some Functional Sites).
- ConSurf Server:
- Results can be visualized and analyzed in FirstGlance in Jmol, which has many advantages.
- You submit proteins of interest and wait for the analysis to be completed.
- Completely automated analysis typically gives excellent results.
- Optionally, highly flexible with many configurable parameters and several sequence database options.
- You can upload your own multiple sequence alignment, or phylogenetic tree, for use in the analysis.
Both servers use state-of-the-art methods that are published in peer-reviewed journals. For comparisons with other methods, see Other Evolutionary Conservation Servers.
Both servers permit you to download results. This is a good idea since the continual growth of sequence databases and improvements in analysis algorithms will give at least slightly different results for the same jobs run several months or more apart. Also, results are periodically deleted from the ConSurf server to conserve disk space.
Examining Functions of Proteins in ConSurf-DB's MSA
- Since 2022, ConSurfDB results are NOT compatible with FirstGlance in Jmol. Visualizing and analyzing conservation results in FirstGlance has many advantages. Use the ConSurf Server for compatibility with FirstGlance.
- In July, 2024, ConSurfDB had not been updated with new entries in the Protein Data Bank since mid-2022. To look for more recent updates, at ConSurfDB Home, click More.
As explained above, ConSurf-DB typically includes proteins with more than one function in its conservation analysis. Before deciding whether to do a ConSurf Server job that limits the analysis to proteins of a single function, you may want to see what proteins ConSurf-DB included in its analysis. Here is how to see the names (which hopefully reveal the functions) of the proteins included in ConSurf-DB's analysis of a protein chain. (The following steps are based on ConSurfDB as of July, 2024.)
- Go to consurfdb.tau.ac.il (the DB, distinct from the ConSurf Server).
- Enter the PDB code (PDB ID) for the protein of interest. If you get "ERROR: No chains found" it means that this PDB entry was released after the most recent update of ConSurfDB, so you should use the ConSurf Server.
- Select the chain of interest. If you're not sure, get familiar with the structure using FirstGlance.
- Click the button Apply, and wait for the results to load.
- Scroll down and click Homologues, Alignment, and Phylogeny.
- Notice the number of sequences in the MSA (the number of "hits" upon which "calculations were conducted").
- To view the list of sequences in the multiple sequence alignment (MSA) from which the conservation pattern was determined, click on the count of hits upon which the calculations were conducted.
- Get the Average Pairwise Distance (APD) by clicking "View Alignment Details". A value close to, or larger than 1.0 suggests that proteins with multiple functions were included in the multiple sequence alignment, which obscures conservation related to the function of your query protein. See Interpreting ConSurf Results, where the APD is explained.
If the list includes proteins of functions that differ from that of the protein of interest, that tends to obscure patches of conservation that exist among proteins with the same function as the query protein of interest.
Example With Multiple Functions
2vaa is a major histocompatibility complex class I protein (MHC I). ConSurfDB used 300 sequences in its calculation, and the MSA had an APD of 1.63. Starting about halfway down the list of sequence is the first MHC II protein, which has a different function. Early in the list is the first of 14 sequences for hemochromatosis proteins (unrelated in function). The list includes 62 "Ig-like domain-containing" proteins of unknown function. The inclusion of these and many other sequences for proteins with functions that differ from the query will obscure conservation of residues critical for the specific functions of MHC I proteins. Therefore, you may prefer to run a ConSurf job in which you limit the APD to a value below 1.0. See such examples below.
Limiting ConSurf Analysis to Proteins of a Single Function
This section was updated in July, 2024 to correspond to changes in the ConSurf Server.
As explained above, ConSurfDB results, and the ConSurf-DB Evolutionary Conservation scene available in Proteopedia often includes proteins with multiple functions. However, the best way to find all functional sites by conservation analysis is to limit the analysis to proteins with a single function.
When the Average Pairwise Distance (APD) in the multiple sequence alignment (MSA) approaches or exceeds approximately 1.0, it is likely that proteins with multiple functions have been included in the MSA. To see conservation that reflects the function of the query protein, it is best to use an MSA with an APD in roughly the range 0.3-0.6. Sometimes, the ConSurf Server's result with default settings may give such a result. If not, you may wish to do additional ConSurf runs with the goal of reducing the APD.
Prior to 2022, the ConSurf Server enabled manual selection of sequences. Unfortunately, after a 2022 update to the ConSurf Server, this is no longer practical. Hence we are limited to adjusting run parameters by trial and error "in the dark" until satisfactory results are obtained.
- Go to consurf.tau.ac.il, the ConSurf Server (distinct from ConSurf-DB).
- Enter your PDB ID in the slot (or upload your PDB file).
- IGNORE the "pre-calculated ConSurfDB analysis" if offered. (Presumably you've already decided it has an APD value too high, and it will not work in FirstGlance in Jmol.)
- Enter a title. Best if the title includes abbreviations for the custom settings you plan for this run. For example, if you are running 2vaa chain A with the UniRef90 database, 150 sequences sampled from the unique hits, with maximal %ID 99% and minimal %ID 60%, the title might be "2vaa u90 150s 99-60".
- Enter your email address.
- Click Select Run Parameters Manually.
Below are suggestions for selecting the run parameters.
APD Is Too High
As explained above and under Interpreting ConSurf Results, when the APD approaches or exceeds 1.0, conservation related to the function of the query protein may be obscured due to inclusion in the MSA of proteins with multiple functions. There are several ways to get less diversity in the MSA:
- Increase the "Minimal %ID" from the default 35% to perhaps 60%-75%. This will exclude the more distantly related sequences.
- Increase the "Maximal %ID" from the default 95% to perhaps 98%. This will include more sequences closely related to the query.
The above two actions are usually sufficient. Actions with larger effects are:
- The default is to choose the 150 sequences in the MSA by uniform sampling of the unique hits. This sample includes both the closest and farthest sequences from the query. Change "sampling" to closest to the unique hits. This typically makes a large drop in the APD. You could reduce the drop by increasing the number of sequences used from 150 to perhaps 250. The more sequences you use, the longer the job will take to complete.
- Try getting sequences from SwissProt instead of UniRef90. SwissProt has roughly 100-fold fewer sequences than does UniRef90. This tends to reduce the APD, but you may have difficulty getting enough sequences to avoid a large number of residues with "insufficient data" (colored yellow).
APD Is Too Low
When the APD drops close to, or falls below, about 0.2, conservation will be exaggerated because of limited diversity in the MSA. You could increase diversity:
- Decrease the "Maximal %ID" below the default of 95%, perhaps to 80% or 60%. This will exclude the most closely related sequences.
- Search a larger database of sequences. UniProt has more sequences than the default of UniRef90, although the additional sequences may not add much diversity. The NR (Non-Redundant) database is the largest (twice as many sequences as UniProt), but contains more errors.
Too Few Sequences
If the default search for sequence homologs fails to find the minimum of 5 sequences, or if you have more than a few yellow residues (yellow means insufficient data to assign a meaningful conservation value):
- Under "Choose parameters for homolog search algorithm", change the Protein Database to UniProt or NR (larger databases than the default Uniref90).
- Increase the "Maximal %ID between sequences" from the default of 95% to perhaps 98%.
If the larger database does not give enough sequences, you can use other options to widen the search for sequences, knowing that you will be retrieving sequences less related to the query sequence, likely including proteins with functions differing from that of the query:
- Increase the number of iterations in the search to more than the default of one. Each iteration generates a sequence profile that is used as a query for the next iteration.
- Increase the default E cutoff of 0.0001, for example, to 0.001 or 0.01.
Too Many Yellow Residues
If more than a few residues are yellow, it means that the MSA had insufficient data to assign meaningful conservation values to the yellow residues. Try to increase the number of sequences in the MSA: See #Too Few Sequences.
If an entire domain of your query protein is yellow (insufficient data) , it is because the multiple sequence alignment (MSA) has poor coverage of the yellow domain. In this case, it is best to do separate ConSurf runs for each domain in your protein.
- Determine sequence numbers for the linkers between domains. This can be done easily by inspection in FirstGlance in Jmol.
- Using a plain text editor, delete everything in your PDB file except the lines that begin with ATOM or HETATM. (Its OK to leave the lines ANISOU if present.)
- Based on sequence numbers, separate the domains into different PDB files.
- Upload each domain's PDB file to ConSurf as a separate job.
Examples
| |||||||||||
Conclusion
In order to discover key functional residues, it is important to inspect multiple ConSurf Server jobs for highly conserved residues, including multiple jobs with #Average Pairwise Distances (APD) in the range 0.25-0.5 using the above methods. Jobs with APD higher than 0.5 may obscure conservation of residues crucial for the function of the query protein. Residues conserved in the broader family of more distantly related proteins with different functions will typically be revealed with default ConSurf Server settings (APD ~ 1.0), or even in the ConSurfDB result.
The ConSurf-DB Mechanism
- In January 2018: ConSurfDB had not been updated with new entries in the Protein Data Bank since January, 2013.
Because results from the ConSurf DataBase server, ConSurf-DB[11] are displayed within Proteopedia as Evolutionary Conservation, an overview of its methods is provided here. ConSurf-DB pre-calculates conservation levels for each amino acid in every protein chain in the Protein Data Bank. It went into service in 2008. It uses state-of-the-art methods, all published in peer-reviewed journals[11].
ConSurf-DB Process
- A list of unique protein chains is extracted from the Protein Data Bank. Chains shorter than 30 amino acids are not processed because they do not contain enough information for reliable phylogenetic tree construction. Certain non-standard residues are converted to the closest standard amino acids, for example, selenomethionine MSE is converted to MET. Chains that still have more than 15% non-standard residues are not processed. Chains that could not be processed are colored gray in Proteopedia -- see the color key at the top of this page.
- The amino acid sequence of each protein chain is submitted to HMMER for collection of related sequences from the UniRef90 database. By default, one iteration is performed using an expectation value[7] cutoff of 10-4.
- The found sequences are then filtered (see below) using a scheme that attempts a balance between limiting the sequences to close homologues, and including distant sequences that do not share structure or function.
- The filtered sequence set is multiply aligned with MAFFT (a multiple sequence alignment algorithm that out-performs older algorithms such as MUSCLE and CLUSTALW).
- A phylogenetic tree is constructed from the multiple sequence alignment (MSA) using the Rate4Site program developed by the ConSurf team.
- Rate4Site then calculates an evolutionary rate for each position in the MSA using a Bayesian approach shown by the ConSurf team to be superior[12]. "The amino acid evolution is traced using the JTT[13] substitution model. High evolutionary rate represents a variable position while low rate represents an evolutionarily conserved position."[11]
- "The conservation scores are normalized so that the average over all residues is zero, and the standard deviation is one."[11] Thus, conservation scores are relative, not absolute and comparing them between different protein families might be misleading (see Caveat above).
- The normalized conservation scores are then divided into nine levels from 1 (highly variable) to 9 (highly conserved).
- Colors mapped to the nine conservation levels, from turquoise (1) to burgandy (9) are applied to the 3D protein structure visualized in FirstGlance in Jmol. A coloring script for RasMol is also provided.

- A confidence interval for the conservation level is calculated for each amino acid position in the MSA. When this indicates low reliability, the position is colored yellow, signifying that the data were insufficient to assign a meaningful conservation level.
- An Average Pairwise Distance (APD) is calculated to describe the diversity of sequences in the MSA (see below).
The results of each stage of the above process may be viewed for each chain at ConSurf-DB. In the initial run (February 2008), roughly 100 computer CPU's were utilized concurrently via a distributed computing system. Processing of the 30,918 unique protein chains in the PDB took about five days, or an average of roughly 30 minutes per chain.
Filtering
Filtering of the sequences gathered for each protein chain is crucial to making the ConSurfDB results maximally informative. Filtering consists of the following steps.
- Sequences with more than 95% sequence identity to the query sequence are discarded.
- Sequences shorter than 60% of the query sequence are discarded.
- Locally aligned sequence fragments that overlap by over 10% are discarded.
- Redundant sequences (>95% identical) are removed using CD-HIT[14].
- A maximum of 300 sequences meeting the above criteria is used (the 300 with the lowest expectation values[7], that is, most closely related to the query sequence).
- If the above process yields fewer than 5 sequence homologs, no calculation is performed due to insufficient data. In February, 2008, this occurred for 1,348 chains out of 30,918 (4%).
Average Pairwise Distance
The Average Pairwise Distance (APD) is an important measure of the diversity in the multiple sequence alignment. To learn how to interpret it, please see above, and Interpreting ConSurf Results.
The ConSurf Server
The ConSurf Server, first available in 2001[15][16][17] with many subsequent enhancements, can calculate and display the conservation pattern for 3D structures completely automatically. It should be used whenever the pre-calculated result at the ConSurf-DB is unavailable, or does not meet your needs (for example, see above), or if you have your own multiple sequence alignment (MSA) that you wish to use. The default settings of ConSurf may need to be adjusted in order to get an optimally informative result. The main adjustment needed is to gather an adequate number of sequences for proteins of the same function as your protein of interest (see above).
ConSurf Server job submission instructions.
Like ConSurf-DB, the ConSurf Server uses the same state-of-the-art methods, all of which are published in peer-reviewed journal articles. Unlike ConSurf-DB's pre-calculated results the ConSurf Server permits considerable customization. For example, the user may specify the number of sequences to use, choose the database from which sequences are obtained, set the Expectation cutoff[7], set the number of HMMER iterations, or submit their own multiple sequence alignment, or phylogenetic tree. Also you can upload your own PDB file, which enables you to process unpublished data, theoretical models, or "trimmed" chains, e.g. a domain of interest from a multiple-domain chain.
In brief, the ConSurf Server uses the following process by default:
- Obtains the protein sequence for the specified PDB code (or uploaded PDB file) and chain.
- Gathers closely related sequences from UNIREF90 (or another database that you specify) with an HMMER search (or other algorithm that you specify). E value cutoff[7], number of iterations, and number of sequences to use are configurable.
- Filters the sequences, by default eliminating those redundant at 95% or higher identity with each other, and those with less than 35% sequence identity to the query sequence. These percentages are adjustable.
- Does a multiple sequence alignment with MAFFT. (Or you can choose a different algorithm or upload your own MSA.)
- Constructs a phylogenetic tree using neighbor joining with ML distance. (Or you can choose a different algorithm or upload your own tree.)
- Calculates a conservation score with confidence interval for each amino acid. Classifies the conservation scores into nine levels, and maps them to standard conservation level colors (see color key at the top of this page). Marks residues for which the conservation score confidence interval is too large, hence the conservation score is unreliable ("insufficient data").
- Displays the protein, colored by conservation, in interactive 3D, using the NGL Viewer, FirstGlance in Jmol, Chimera, or PyMOL.
See Also
- ConSurf/Index provides links to all pages about evolutionary conservation and ConSurf in Proteopedia.
Notes & References
- ↑ Tested with 20 arbitrarily selected proteins, mostly enzymes. Average of the average pairwise distance (APD) values: 1.00; range 0.82-1.42.
- ↑ Tested with 20 arbitrarily selected proteins, mostly enzymes. Average number of amino acids with insufficient data ("yellow" in ConSurf): 3.5; range 0 to 16.
- ↑ 3.0 3.1 Default ConSurf Server settings: UniRef90 database, excluding sequences with > 95% or < 35% identity with the query, MSA has 150 sequences sampled evenly from all unique sequence hits.
- ↑ Custom ConSurf Server settings for APD 0.51 with 2vaa: UniRef90 database, excluding sequences with > 95% or < 50% identity with the query, MSA has 150 sequences sampled evenly from all unique sequence hits.
- ↑ Custom ConSurf Server settings for APD 0.31 with 2vaa: UniRef90 database, excluding sequences with > 98% or < 70% identity with the query, MSA has 150 sequences sampled evenly from all unique sequence hits.
- ↑ Custom ConSurf Server settings for APD 0.30 with 2vaa: UniRef90 database, excluding sequences with > 95% or < 35% identity with the query, MSA has 250 sequences closest to the query.
- ↑ 7.0 7.1 7.2 7.3 7.4 Expectation Value (E value): When searching a sequence database with a query sequence, e.g. using BLAST or PSI-BLAST, each found sequence can be characterized by an E value. It is the number of hits expected by chance with the sequence matching level observed, taking into account the size of the sequence database and length of the query sequence. Low values of E (much less than one) mean increasing significance of the match.
- ↑ 8.0 8.1 With APD 0.48, mean conservation of between-chain salt bridged atoms is 7.57 ± 0.13 SEM. Subtracting 3 SEM (99% confidence limit) gives 7.18. This does not overlap with either 7.16 (the all-salt-bridged atoms mean + 3 SEM) or 6.82 (the mean for within-chain salt-bridged atoms + 3 SEM).
- ↑ ConSurf settings for APD 0.91 with 4dnw: Clean UniProt, 35-95%, 200 sequences closest to query.
- ↑ ConSurf settings for APD 0.48 with 4dnw: Clean UniProt, 35-95%, 125 sequences closest to query.
- ↑ 11.0 11.1 11.2 11.3 Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009 Jan;37(Database issue):D323-7. Epub 2008 Oct 29. PMID:18971256 doi:http://dx.doi.org/10.1093/nar/gkn822
- ↑ Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol Biol Evol. 2004 Sep;21(9):1781-91. Epub 2004 Jun 16. PMID:15201400 doi:http://dx.doi.org/10.1093/molbev/msh194
- ↑ Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992 Jun;8(3):275-82. PMID:1633570
- ↑ Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006 Jul 1;22(13):1658-9. Epub 2006 May 26. PMID:16731699 doi:http://dx.doi.org/10.1093/bioinformatics/btl158
- ↑ Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001 Mar 16;307(1):447-63. PMID:11243830 doi:http://dx.doi.org/10.1006/jmbi.2000.4474
- ↑ Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003 Jan;19(1):163-4. PMID:12499312
- ↑ Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W299-302. PMID:15980475 doi:http://dx.doi.org/33/suppl_2/W299